Amyloid fibril formation by an SH3 domain
- PMID: 9539718
- PMCID: PMC22470
- DOI: 10.1073/pnas.95.8.4224
Amyloid fibril formation by an SH3 domain
Abstract
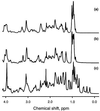
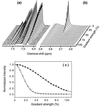
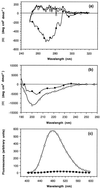
The SH3 domain is a well characterized small protein module with a simple fold found in many proteins. At acid pH, the SH3 domain (PI3-SH3) of the p85alpha subunit of bovine phosphatidylinositol 3-kinase slowly forms a gel that consists of typical amyloid fibrils as assessed by electron microscopy, a Congo red binding assay, and x-ray fiber diffraction. The soluble form of PI3-SH3 at acid pH (the A state by a variety of techniques) from which fibrils are generated has been characterized. Circular dichroism in the far- and near-UV regions and 1H NMR indicate that the A state is substantially unfolded relative to the native protein at neutral pH. NMR diffusion measurements indicate, however, that the effective hydrodynamic radius of the A state is only 23% higher than that of the native protein and is 20% lower than that of the protein denatured in 3.5 M guanidinium chloride. In addition, the A state binds the hydrophobic dye 1-anilinonaphthalene-8-sulfonic acid, which suggests that SH3 in this state has a partially formed hydrophobic core. These results indicate that the A state is partially folded and support the hypothesis that partially folded states formed in solution are precursors of amyloid deposition. Moreover, that this domain aggregates into amyloid fibrils suggests that the potential for amyloid deposition may be a common property of proteins, and not only of a few proteins associated with disease.
Figures


References
-
- Tan S Y, Pepys M B. Histopathology. 1994;25:403–414. - PubMed
-
- Kelly J W. Curr Op Struct Biol. 1996;6:11–17. - PubMed
-
- Sunde M, Blake C C F. Adv Protein Chem. 1997;50:123–159. - PubMed
-
- Blake C, Serpell L C. Structure. 1996;4:989–998. - PubMed
-
- Sunde M, Serpell L C, Bartlam M, Fraser P E, Pepys M B, Blake C C F. J Mol Biol. 1997;273:729–739. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

