Polymerase recognition of synthetic oligodeoxyribonucleotides incorporating degenerate pyrimidine and purine bases
- PMID: 9539724
- PMCID: PMC22476
- DOI: 10.1073/pnas.95.8.4258
Polymerase recognition of synthetic oligodeoxyribonucleotides incorporating degenerate pyrimidine and purine bases
Abstract
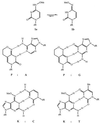
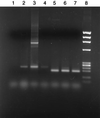
A universal base that is capable of substituting for any of the four natural bases in DNA would be of great utility in both mutagenesis and recombinant DNA experiments. This paper describes the properties of oligonucleotides incorporating two degenerate bases, the pyrimidine base 6H,8H-3,4-dihydropyrimido[4,5-c][1,2]oxazin-7-one and the purine base N6-methoxy-2,6-diaminopurine, designated P and K, respectively. An equimolar mixture of the analogues P and K (called M) acts, in primers, as a universal base. The thermal stability of oligonucleotide duplexes were only slightly reduced when natural bases were replaced by P or K. Templates containing the modified bases were copied by Taq polymerase; P behaved as thymine in 60% of copying events and as cytosine in 40%, whereas K behaved as if it were guanine (13%) or adenine (87%). The dUTPase gene of Caenorhabditis elegans, which we have found to contain three nonidentical homologous repeats, was used as a model system to test the use of these bases in primers for DNA synthesis. A pair of oligodeoxyribonucleotides, each 20 residues long and containing an equimolar mixture of P and K at six positions, primed with high specificity both T7 DNA polymerase in sequencing reactions and Taq polymerase in PCRs; no nonspecific amplification was obtained on genomic DNA of C. elegans. Use of P and K can significantly reduce the complexity of degenerate oligonucleotide mixtures, and when used together, P and K can act as a universal base.
Figures




Similar articles
-
Biochemical behavior of N-oxidized cytosine and adenine bases in DNA polymerase-mediated primer extension reactions.Nucleic Acids Res. 2011 Apr;39(7):2995-3004. doi: 10.1093/nar/gkq914. Epub 2011 Feb 7. Nucleic Acids Res. 2011. PMID: 21300642 Free PMC article.
-
Synthesis of oligodeoxyribonucleotides containing degenerate bases and their use as primers in the polymerase chain reaction.Nucleic Acids Res. 1992 Oct 11;20(19):5149-52. doi: 10.1093/nar/20.19.5149. Nucleic Acids Res. 1992. PMID: 1408830 Free PMC article.
-
15-mer DNA duplexes containing an abasic site are thermodynamically more stable with adjacent purines than with pyrimidines.Biochemistry. 2001 Apr 3;40(13):3859-68. doi: 10.1021/bi0024409. Biochemistry. 2001. PMID: 11300765
-
The structure and application of oligodeoxyribonucleotides containing modified, degenerate bases.Nucleic Acids Symp Ser. 1991;(24):209-12. Nucleic Acids Symp Ser. 1991. PMID: 1841286
-
Redesigning the architecture of the base pair: toward biochemical and biological function of new genetic sets.Chem Biol. 2009 Mar 27;16(3):242-8. doi: 10.1016/j.chembiol.2008.12.004. Chem Biol. 2009. PMID: 19318205 Free PMC article. Review.
Cited by
-
Introduction of mismatches in a random shRNA-encoding library improves potency for phenotypic selection.PLoS One. 2014 Feb 3;9(2):e87390. doi: 10.1371/journal.pone.0087390. eCollection 2014. PLoS One. 2014. PMID: 24498319 Free PMC article.
-
Lethal mutagenesis of poliovirus mediated by a mutagenic pyrimidine analogue.J Virol. 2007 Oct;81(20):11256-66. doi: 10.1128/JVI.01028-07. Epub 2007 Aug 8. J Virol. 2007. PMID: 17686844 Free PMC article.
-
DENT-seq for genome-wide strand-specific identification of DNA single-strand break sites with single-nucleotide resolution.Genome Res. 2021 Jan;31(1):75-87. doi: 10.1101/gr.265223.120. Epub 2020 Dec 21. Genome Res. 2021. PMID: 33355294 Free PMC article.
-
Saturation of DNA mismatch repair and error catastrophe by a base analogue in Escherichia coli.Genetics. 2002 Aug;161(4):1363-71. doi: 10.1093/genetics/161.4.1363. Genetics. 2002. PMID: 12196386 Free PMC article.
-
Quantitative multiplex assay for simultaneous detection and identification of Indiana and New Jersey serotypes of vesicular stomatitis virus.J Clin Microbiol. 2005 Jan;43(1):356-62. doi: 10.1128/JCM.43.1.356-362.2005. J Clin Microbiol. 2005. PMID: 15634994 Free PMC article.
References
-
- Ohtsuka E, Matsuki S, Ikehara M, Takahashi Y, Matsubara K. J Biol Chem. 1985;260:2605–2608. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

