Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia
- PMID: 9539781
- PMCID: PMC22533
- DOI: 10.1073/pnas.95.8.4584
Fetal origins of the TEL-AML1 fusion gene in identical twins with leukemia
Abstract
The TEL (ETV6)-AML1 (CBFA2) gene fusion is the most common reciprocal chromosomal rearrangement in childhood cancer occurring in approximately 25% of the most predominant subtype of leukemia- common acute lymphoblastic leukemia. The TEL-AML1 genomic sequence has been characterized in a pair of monozygotic twins diagnosed at ages 3 years, 6 months and 4 years, 10 months with common acute lymphoblastic leukemia. The twin leukemic DNA shared the same unique (or clonotypic) but nonconstitutive TEL-AML1 fusion sequence. The most plausible explanation for this finding is a single cell origin of the TEL-AML fusion in one fetus in utero, probably as a leukemia-initiating mutation, followed by intraplacental metastasis of clonal progeny to the other twin. Clonal identity is further supported by the finding that the leukemic cells in the two twins shared an identical rearranged IGH allele. These data have implications for the etiology and natural history of childhood leukemia.
Figures
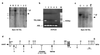



References
-
- Sawyers C L. Lancet. 1997;349:196–200. - PubMed
-
- Hagemeijer A, Grosveld G. In: Leukemia. Henderson E S, Lister T A, Greaves M F, editors. Philadelphia: Saunders; 1996. pp. 131–144.
-
- Rabbitts T H. Nature (London) 1994;372:143–149. - PubMed
-
- Look A T. Science. 1997;278:1059–1064. - PubMed
-
- Golub T R, Barker G F, Stegmaier K, Gilliland D G. Curr Top Microbiol Immunol. 1996;211:279–288. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

