Characterization of the rrnB operon of the plant pathogen Rhodococcus fascians and targeted integrations of exogenous genes at rrn loci
- PMID: 9546162
- PMCID: PMC106141
- DOI: 10.1128/AEM.64.4.1276-1282.1998
Characterization of the rrnB operon of the plant pathogen Rhodococcus fascians and targeted integrations of exogenous genes at rrn loci
Abstract
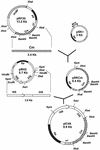
A 6.0-kb SalI DNA fragment containing an entire rRNA operon (rrnB) was cloned from a cosmid gene bank of the phytopathogenic strain Rhodococcus fascians D188. The nucleotide sequence of the 6-kb fragment was determined and had the organization 16S rRNA-spacer-23S rRNA-spacer-5S rRNA without tRNA-encoding genes in the spacer regions. The 5' and 3' ends of the mature 16S, 23S, and 5S rRNAs were determined by alignment with the rrn operons of Bacillus subtilis and other gram-positive bacteria. Four copies of the rrn operons were identified by hybridization with an rrnB probe in R. fascians type strain ATCC 12974 and in the virulent strain R. fascians D188. However, another isolate, CECT 3001 (= NRRL B15096), also classified as R. fascians, produced five rrn-hybridizing bands. An integrative vector containing a 2.5-kb DNA fragment internal to rrnB was constructed for targeted integration of exogenous genes at the rrn loci. Transformants carrying the exogenous chloramphenicol resistance gene (cmr) integrated in different rrn operons were obtained. These transformants had normal growth rates in complex medium and minimal medium and were fully stable for the integrated marker.
Figures




References
-
- Amador, E., J. M. Castro, A. Correia, and J. F. Martín. Unpublished data.
-
- Bathe B, Kalinowsky J, Puhler A. A physical and genetic map of the Corynebacterium glutamicum ATCC 13032 chromosome. Mol Gen Genet. 1996;252:255–265. - PubMed
-
- Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

