Lack of intraclonal diversification in Ig heavy and light chain V region genes expressed by CD5+IgM+ chronic lymphocytic leukemia B cells: a multiple time point analysis
- PMID: 9551917
- PMCID: PMC4625536
Lack of intraclonal diversification in Ig heavy and light chain V region genes expressed by CD5+IgM+ chronic lymphocytic leukemia B cells: a multiple time point analysis
Abstract
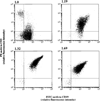
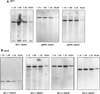
To analyze the modalities of clonal expansion of chronic lymphocytic leukemia (CLL) cells, we sequenced at multiple time points the V(D)J genes expressed by CD5+IgM+CLL B cells in three patients. All three V(D)J gene sequences were found to be point mutated. The mutation frequency in the Ig VH (3.96 x 10(-2) and 2.41 x 10(-2) change/bp) and Vkappa and Vlambda (6.67 x 10(-2) and 1.74 x 10(-2) change/bp) genes of two CLLs (1.19 and 1.32, respectively) was similar, and higher than that in the corresponding gene segments of the third CLL (1.69; 3.4 x 10(-3) and 6.67 x 10(-3) change/bp). In all three CLLs, there was no preferential representation of nucleotide changes yielding amino acid replacement (R mutations), nor was there any preferential segregation of R mutations within the Ig V gene complementarity-determining regions. In all three CLLs, the somatic mutations were all identical in multiple Ig VHDJH transcripts at any given time point, and were all conserved at multiple time points throughout a 2-yr period. The lack of concentration of R mutations in the complementarity-determining regions and the lack of intraclonal heterogeneity suggest that Ag may no longer be able to play a significant role in the clonal expansion of these cells. This conclusion would be strengthened further by the germline configuration of the bcl-1 and bcl-2 proto-oncogenes that are translocated in neoplastic B cells that display significant traces of intraclonal diversification and Ag-dependent selection, such as B-prolymphocytic leukemia and low grade follicular non-Hodgkin lymphoma.
Figures


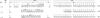

References
-
- Zukerberg LR, Jeffrey L, Medeiros L, Ferry JA, Harris NL. Diffuse low-grade B-cell lymphomas. Am. J. Clin. Pathol. 1993;100:373. - PubMed
-
- Newman RA, Peterson B, Davey FR, Brabyn C, Collins H, Brunetto VL, Duggan DB, Weiss RB, Royston I, Millard FE, Miller AA, Bloomfield CD. Phenotypic markers and bcl-1 gene rearrangements in B-cell chronic lymphocytic leukemia: a cancer and leukemia group B study. Blood. 1993;82:1239. - PubMed
-
- Mayer R, Logtenberg T, Strauchen J, Dimitriu-Bona A, Mayer L, Mechanic S, Chiorazzi N, Borche L, Dighiero G, Mannheimer-Lory A, Diamond B, Alt F, Bona C. CD5 and immunoglobulin V gene expression in B-cell lymphomas and chronic lymphocytic leukemia. Blood. 1990;75:1518. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
