Identification of the omega4400 regulatory region, a developmental promoter of Myxococcus xanthus
- PMID: 9555878
- PMCID: PMC107122
- DOI: 10.1128/JB.180.8.1995-2004.1998
Identification of the omega4400 regulatory region, a developmental promoter of Myxococcus xanthus
Abstract
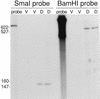
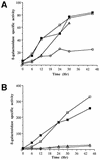
Omega4400 is the site of a Tn5 lac insertion in the Myxococcus xanthus genome that fuses lacZ expression to a developmentally regulated promoter. Cell-cell interactions that occur during development, including C signaling, are required for normal expression of Tn5 lac omega4400. The DNA upstream of the omega4400 insertion has been cloned, the promoter has been localized, and a partial open reading frame has been identified. From the deduced amino acid sequence of the partial open reading frame, the gene disrupted by Tn5 lac omega4400 may encode a protein with an ATP- or GTP-binding site. Expression of the gene begins 6 to 12 h after starvation initiates development, as measured by beta-galactosidase production in cells containing Tn5 lac omega4400. The putative transcriptional start site was mapped, and deletion analysis has shown that DNA downstream of -101 bp is sufficient for C-signal-dependent, developmental activation of this promoter. A deletion to -76 bp eliminated promoter activity, suggesting the involvement of an upstream activator protein. The promoter may be transcribed by RNA polymerase containing a novel sigma factor, since a mutation in the M. xanthus sigB or sigC gene did not affect Tn5 lac omega4400 expression and the DNA sequence upstream of the transcriptional start site did not match the sequence of any M. xanthus promoter transcribed by a known form of RNA polymerase. However, the omega4400 promoter does contain the sequence 5'-CATCCCT-3' centered at -49 and the C-signal-dependent omega4403 promoter also contains this sequence at the same position. Moreover, the two promoters match at five of six positions in the -10 regions, suggesting that these promoters may share one or more transcription factors. These results begin to define the cis-acting regulatory elements important for cell-cell interaction-dependent gene expression during the development of a multicellular prokaryote.
Figures






References
-
- Apelian D, Inouye S. Development-specific ς-factor essential for late-stage differentiation of Myxococcus xanthus. Genes Dev. 1990;4:1396–1403. - PubMed
-
- Avery L, Kaiser D. In situ transposon replacement and isolation of a spontaneous tandem genetic duplication. Mol Gen Genet. 1983;191:99–109. - PubMed
-
- Biran, D., and L. Kroos. Unpublished data.
-
- Biran D, Kroos L. In vitro transcription of Myxococcus xanthus genes with RNA polymerase containing ςA, the major sigma factor in growing cells. Mol Microbiol. 1997;25:463–472. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

