The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by sigmaF, and KatX is essential for hydrogen peroxide resistance of the germinating spore
- PMID: 9555886
- PMCID: PMC107130
- DOI: 10.1128/JB.180.8.2057-2062.1998
The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by sigmaF, and KatX is essential for hydrogen peroxide resistance of the germinating spore
Abstract
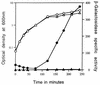
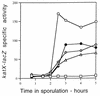
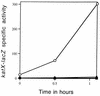
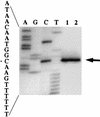
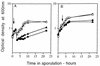
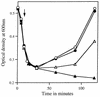
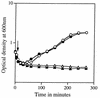
Previous work has shown that the katX gene encodes the major catalase in dormant spores of Bacillus subtilis but that this enzyme has no role in dormant spore resistance to hydrogen peroxide. Expression of a katX-lacZ fusion began at approximately h 2 of sporulation, and >75% of the katX-driven beta-galactosidase was packaged into the mature spore. A mutation in the gene coding for the sporulation-specific RNA polymerase sigma factor sigmaF abolished katX-lacZ expression, while mutations in genes encoding sigmaE, sigmaG, and sigmaK did not. Induction of sigmaF synthesis in vegetative cells also resulted in katX-lacZ expression, while induction of sigmaG expression did not; the katX-lacZ fusion was also not induced by hydrogen peroxide. Upstream of the in vivo katX transcription start site there are sequences with good homology to those upstream of known sigmaF-dependent start sites. These data indicate that katX is an additional member of the forespore-specific sigmaF regulon. A mutant in the katA gene, encoding the major catalase in growing cells, was sensitive to hydrogen peroxide during sporulation, while a katX mutant was not. However, outgrowth of katX spores, but not katA spores, was sensitive to hydrogen peroxide. Consequently, a major function for KatX is to protect germinating spores from hydrogen peroxide.
Figures


References
-
- Alper S, Duncan L, Losick R. An adenosine nucleotide switch controlling the activity of a cell type-specific transcription factor in B. subtilis. Cell. 1994;77:195–205. - PubMed
-
- Bol D K, Yasbin R E. The isolation, cloning and identification of a vegetative catalase gene from Bacillus subtilis. Gene. 1991;109:31–37. - PubMed
-
- Casillas-Martinez, L., and P. Setlow. 1997. Unpublished results.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

