Identification and characterization of epoxide carboxylase activity in cell extracts of Nocardia corallina B276
- PMID: 9555888
- PMCID: PMC107132
- DOI: 10.1128/JB.180.8.2072-2078.1998
Identification and characterization of epoxide carboxylase activity in cell extracts of Nocardia corallina B276
Abstract
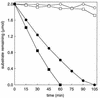
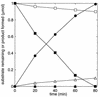
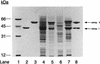
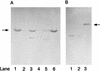
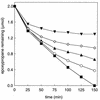
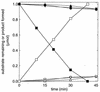
The metabolism of aliphatic epoxides (epoxyalkanes) by the alkene-utilizing actinomycete Nocardia corallina B276 was investigated. Suspensions of N. corallina cells grown with propylene as the carbon source readily degraded propylene and epoxypropane, while suspensions of glucose-grown cells did not. The addition of propylene and epoxypropane to glucose-grown cells resulted in a time-dependent increase in propylene- and epoxypropane-degrading activities that was prevented by the addition of rifampin and chloramphenicol. The expression of alkene- and epoxide-degrading activities was correlated with the high-level expression of several polypeptides not present in extracts of glucose-grown cells. Epoxypropane and epoxybutane degradation by propylene-grown cell suspensions of N. corallina was stimulated by the addition of CO2 and inhibited by the depletion of CO2. Cell extracts catalyzed the carboxylation of epoxypropane to form acetoacetate in a reaction that was dependent on the addition of CO2, NAD+, and a reductant (NADPH or dithiothreitol). In the absence of CO2, epoxypropane was isomerized by cell extracts to form acetone at a rate approximately 10-fold lower than the rate of epoxypropane carboxylation. Methylepoxypropane was found to be a time-dependent, irreversible inactivator of epoxyalkane-degrading activity. These properties demonstrate that epoxyalkane metabolism in N. corallina occurs by a carboxylation reaction forming beta-keto acids as products and provide evidence for the involvement in this reaction of an epoxide carboxylase with properties and cofactor requirements similar to those of the four-component epoxide carboxylase enzyme system of the gram-negative bacterium Xanthobacter strain Py2 (J. R. Allen and S. A. Ensign, J. Biol. Chem. 272:32121-32128, 1997). The addition of epoxide carboxylase component I from Xanthobacter strain Py2 to methylepoxypropane-inactivated N. corallina extracts restored epoxide carboxylase activity, and the addition of epoxide carboxylase component II from Xanthobacter Py2 to active N. corallina extracts stimulated epoxide isomerase rates to the same levels observed with the purified Xanthobacter system. Antibodies raised against Xanthobacter strain Py2 epoxide carboxylase component I cross-reacted with a polypeptide in propylene-grown N. corallina extracts with the same molecular weight as component I but did not cross-react with glucose-grown extracts. Together, these results suggest a common pathway of epoxyalkane metabolism for phylogenetically distinct bacteria that involves CO2 fixation and the activity of a multicomponent epoxide carboxylase enzyme system.
Figures


References
-
- Allen J R, Ensign S A. Purification to homogeneity and reconstitution of the individual components of the epoxide carboxylase multiprotein enzyme complex from Xanthobacter strain Py2. J Biol Chem. 1997;272:32121–32128. - PubMed
-
- Chromy V, Fischer J, Kulhanek V. Re-evaluation of EDTA-chelated biuret reagent. Clin Chem. 1974;20:1362–1363. - PubMed
-
- de Bont J A M, van Dijken J P, van Ginkel C G. The metabolism of 1,2-propanediol by the propylene oxide utilizing bacterium Nocardia A60. Biochim Biophys Acta. 1982;714:465–470.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

