A novel DNA polymerase family found in Archaea
- PMID: 9555910
- PMCID: PMC107154
- DOI: 10.1128/JB.180.8.2232-2236.1998
A novel DNA polymerase family found in Archaea
Abstract
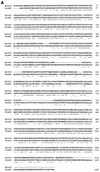
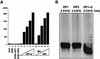
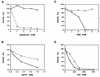
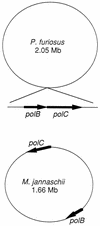
One of the most puzzling results from the complete genome sequence of the methanogenic archaeon Methanococcus jannaschii was that the organism may have only one DNA polymerase gene. This is because no other DNA polymerase-like open reading frames (ORFs) were found besides one ORF having the typical alpha-like DNA polymerase (family B). Recently, we identified the genes of DNA polymerase II (the second DNA polymerase) from the hyperthermophilic archaeon Pyrococcus furiosus, which has also at least one alpha-like DNA polymerase (T. Uemori, Y. Sato, I. Kato, H. Doi, and Y. Ishino, Genes Cells 2:499-512, 1997). The genes in M. jannaschii encoding the proteins that are homologous to the DNA polymerase II of P. furiosus have been located and cloned. The gene products of M. jannaschii expressed in Escherichia coli had both DNA polymerizing and 3'-->5' exonuclease activities. We propose here a novel DNA polymerase family which is entirely different from other hitherto-described DNA polymerases.
Figures


References
-
- Brush G S, Kelly T J. Mechanisms for replicating DNA. In: DePamphilis M L, editor. DNA replication in eukaryotic cells. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1996. pp. 1–43.
-
- Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
-
- Edgell D R, Doolittle W F. Archaebacterial genomics: the complete genome sequence of Methanococcus jannaschii. Bioessays. 1996;19:1–4.
-
- Edgell D R, Doolittle W F. Archaea and the origin(s) of DNA replication proteins. Cell. 1997;89:995–998. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

