The roles of Pol and Env in the assembly pathway of human foamy virus
- PMID: 9557646
- PMCID: PMC109586
- DOI: 10.1128/JVI.72.5.3658-3665.1998
The roles of Pol and Env in the assembly pathway of human foamy virus
Abstract
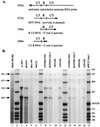
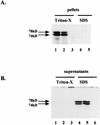
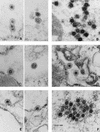
Human foamy virus (HFV) is the prototype of the Spumavirus genus of retroviruses. These viruses have a genomic organization close to that of other complex retroviruses but have similarities to hepadnaviruses such as human hepatitis B virus (HBV). Both HFV and HBV express their Pol protein independently of their structural proteins. Retroviruses and hepadnaviruses differ in their requirements for particle assembly and genome packaging. Assembly of retroviral particles containing RNA genomes requires only the Gag structural protein. The Pol protein is not required for capsid assembly, and the Env surface glycoprotein is not required for release of virions from the cell. In contrast, assembly of extracellular HBV particles containing DNA requires core structural protein and polymerase (P protein) for assembly of nucleocapsids and requires surface glycoproteins for release from the cell. We investigated the requirements for synthesis of extracellular HFV particles by constructing mutants with either the pol or env gene deleted. We found that the Pol protein is dispensable for production of extracellular particles containing viral nucleic acid. In the absence of Env, intracellular particles are synthesized but few or no extracellular particles could be detected. Thus, foamy virus assembly is distinct from that of other reverse transcriptase-encoding mammalian viruses.
Figures
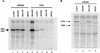
Comment in
-
Endogenous virus of BHK-21 cells complicates electron microscopy studies of foamy virus maturation.J Virol. 1999 Oct;73(10):8917. doi: 10.1128/JVI.73.10.8917-8917.1999. J Virol. 1999. PMID: 10523150 Free PMC article. No abstract available.
References
-
- Achong B G, Mansell W A, Epstein M A, Clifford P. An unusual virus in cultures from a human nasopharyngeal carcinoma. J Natl Cancer Inst. 1971;46:299–307. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

