Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease
- PMID: 9557672
- PMCID: PMC109612
- DOI: 10.1128/JVI.72.5.3872-3886.1998
Simian immunodeficiency virus replicates to high levels in sooty mangabeys without inducing disease
Abstract
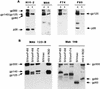
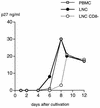
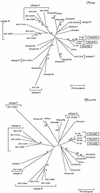
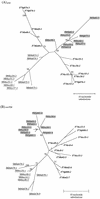
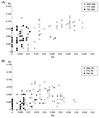
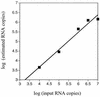
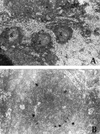
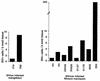
A serologic survey of primates living in a French zoo allowed identification of three cases of infection with simian immunodeficiency virus in sooty mangabeys (Cercocebus atys) (SIVsm). Viral isolates, which were designated SIVsmFr66, SIVsmFr74, and SIVsmFr85, were obtained after short-term culture of mangabey lymphoid cells. Phylogenetic analysis of gag and env sequences amplified directly from mangabey tissues showed that the three SIVsmFr were genetically close and that they constituted a new subtype within the diverse SIVsm-SIVmac-human immunodeficiency virus type 2 (HIV-2) group. We could reconstruct the transmission events that likely occurred in 1986 between the three animals and evaluate the divergence of SIVsmFr sequences since transmission. The estimated rate of mutation fixation was 6 x 10(-3) substitutions per site per year, which was as high as the rate found for SIVmac infection in macaques. These data indicated that SIVsmFr replicated at a high rate in mangabeys, despite the nonpathogenic character of infection in this host. The viral load evaluated by competitive PCR reached 20,000 viral DNA copies per 10(6) lymph node cells. In addition, productively infected cells were readily detected in mangabey lymphoid tissues by in situ hybridization. The amounts of viral RNA in plasma ranged from 10(5) to 10(7) copies per ml. The cell-associated and plasma viral loads were as high as those seen in susceptible hosts (humans or macaques) during the asymptomatic stage of HIV or SIVmac infections. Thus, the lack of pathogenicity of SIVsm for its natural host cannot be explained by limited viral replication or by tight containment of viral production.
Figures


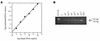
References
-
- Ahmed-Ansari A, Powell J D, Jensen P E, Yehuda-Cohen T, McClure H M, Anderson D, Fultz P N, Sell K W. Requirement for simian immunodeficiency virus antigen-specific in vitro proliferation of T-cells from infected rhesus macaques and sooty mangabeys. AIDS. 1990;4:399–407. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

