Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid
- PMID: 9557680
- PMCID: PMC109620
- DOI: 10.1128/JVI.72.5.3944-3951.1998
Assembly of the herpes simplex virus capsid: preformed triplexes bind to the nascent capsid
Abstract
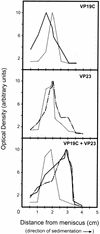
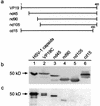
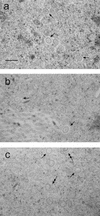
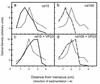
The herpes simplex virus type 1 (HSV-1) capsid is a T=16 icosahedral shell that forms in the nuclei of infected cells. Capsid assembly also occurs in vitro in reaction mixtures created from insect cell extracts containing recombinant baculovirus-expressed HSV-1 capsid proteins. During capsid formation, the major capsid protein, VP5, and the scaffolding protein, pre-VP22a, condense to form structures that are extended into procapsids by addition of the triplex proteins, VP19C and VP23. We investigated whether triplex proteins bind to the major capsid-scaffold protein complexes as separate polypeptides or as preformed triplexes. Assembly products from reactions lacking one triplex protein were immunoprecipitated and examined for the presence of the other. The results showed that neither triplex protein bound unless both were present, suggesting that interaction between VP19C and VP23 is required before either protein can participate in the assembly process. Sucrose density gradient analysis was employed to determine the sedimentation coefficients of VP19C, VP23, and VP19C-VP23 complexes. The results showed that the two proteins formed a complex with a sedimentation coefficient of 7.2S, a value that is consistent with formation of a VP19C-VP23(2) heterotrimer. Furthermore, VP23 was observed to have a sedimentation coefficient of 4.9S, suggesting that this protein exists as a dimer in solution. Deletion analysis of VP19C revealed two domains that may be required for attachment of the triplex to major capsid-scaffold protein complexes; none of the deletions disrupted interaction of VP19C with VP23. We propose that preformed triplexes (VP19C-VP23(2) heterotrimers) interact with major capsid-scaffold protein complexes during assembly of the HSV-1 capsid.
Figures


Similar articles
-
Assembly of the herpes simplex virus procapsid from purified components and identification of small complexes containing the major capsid and scaffolding proteins.J Virol. 1999 May;73(5):4239-50. doi: 10.1128/JVI.73.5.4239-4250.1999. J Virol. 1999. PMID: 10196320 Free PMC article.
-
Functional analysis of the triplex proteins (VP19C and VP23) of herpes simplex virus type 1.J Virol. 2006 Jan;80(2):929-40. doi: 10.1128/JVI.80.2.929-940.2006. J Virol. 2006. PMID: 16378995 Free PMC article.
-
A domain in the herpes simplex virus 1 triplex protein VP23 is essential for closure of capsid shells into icosahedral structures.J Virol. 2011 Dec;85(23):12698-707. doi: 10.1128/JVI.05791-11. Epub 2011 Sep 28. J Virol. 2011. PMID: 21957296 Free PMC article.
-
The herpes simplex virus procapsid: structure, conformational changes upon maturation, and roles of the triplex proteins VP19c and VP23 in assembly.J Mol Biol. 1996 Nov 1;263(3):447-62. doi: 10.1016/s0022-2836(96)80018-0. J Mol Biol. 1996. PMID: 8918600
-
Herpesvirus Capsid Assembly and DNA Packaging.Adv Anat Embryol Cell Biol. 2017;223:119-142. doi: 10.1007/978-3-319-53168-7_6. Adv Anat Embryol Cell Biol. 2017. PMID: 28528442 Free PMC article. Review.
Cited by
-
Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization.J Bacteriol. 2005 Mar;187(6):2050-7. doi: 10.1128/JB.187.6.2050-2057.2005. J Bacteriol. 2005. PMID: 15743953 Free PMC article.
-
Herpes simplex virus 1 DNA packaging proteins encoded by UL6, UL15, UL17, UL28, and UL33 are located on the external surface of the viral capsid.J Virol. 2006 Nov;80(21):10894-9. doi: 10.1128/JVI.01364-06. Epub 2006 Aug 18. J Virol. 2006. PMID: 16920825 Free PMC article.
-
The marine algal virus PpV01 has an icosahedral capsid with T=219 quasisymmetry.J Virol. 2005 Jul;79(14):9236-43. doi: 10.1128/JVI.79.14.9236-9243.2005. J Virol. 2005. PMID: 15994818 Free PMC article.
-
Cytomegalovirus capsid protease: biological substrates are cleaved more efficiently by full-length enzyme (pUL80a) than by the catalytic domain (assemblin).J Virol. 2011 Apr;85(7):3526-34. doi: 10.1128/JVI.02663-10. Epub 2011 Jan 26. J Virol. 2011. PMID: 21270147 Free PMC article.
-
Functions of Varicella-zoster virus ORF23 capsid protein in viral replication and the pathogenesis of skin infection.J Virol. 2008 Oct;82(20):10231-46. doi: 10.1128/JVI.01890-07. Epub 2008 Aug 6. J Virol. 2008. PMID: 18684828 Free PMC article.
References
-
- Aebi U, van Driel R, Bijlenga R K, ten Heggeler B, van den Broek R, Steven A C, Smith P R. Capsid fine structure of T-even bacteriophages. Binding and localization of two dispensable capsid proteins into the P23* surface lattice. J Mol Biol. 1977;110:687–698. - PubMed
-
- Baxter M K, Gibson W. 22nd International Herpesvirus Workshop, La Jolla, Calif. 1997. The putative cytomegalovirus triplex proteins, minor capsid protein (mCP) and mCP-binding protein (mC-BP) form a heterotrimeric complex that localizes to the cell nucleus in the absence of other viral proteins, abstr. 168.
-
- Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calender R, editor. The bacteriophages. Vol. 1. New York, N.Y: Plenum Publishing Corp.; 1988. pp. 15–91.
-
- Desai P, Person S. Molecular interactions between the HSV-1 capsid proteins as measured by the yeast two-hybrid system. Virology. 1996;220:516–521. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

