Cloning of novel isoforms of the human Gli2 oncogene and their activities to enhance tax-dependent transcription of the human T-cell leukemia virus type 1 genome
- PMID: 9557682
- PMCID: PMC109622
- DOI: 10.1128/JVI.72.5.3958-3964.1998
Cloning of novel isoforms of the human Gli2 oncogene and their activities to enhance tax-dependent transcription of the human T-cell leukemia virus type 1 genome
Abstract
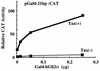
The expression of human T-cell leukemia virus type 1 (HTLV-1) is activated by interaction of a viral transactivator protein, Tax, and cellular transcription factor, CREB (cyclic AMP response element binding protein), which bind to a 21-bp enhancer in the long terminal repeats (LTR). THP (Tax-helping protein) was previously determined to enhance the transactivation by Tax protein. Here we report novel forms of the human homolog of a member of the Gli oncogene family, Gli2 (also termed Gli2/THP), an extended form of a zinc finger protein, THP, which was described previously. Four possible isoforms (hGli2 alpha, beta, gamma, and delta) are formed by combinations of two independent alternative splicings, and all the isoforms could bind to a DNA motif, TRE2S, in the LTR. The longer isoforms, alpha and beta, were abundantly expressed in various cell lines including HTLV-1-infected T-cell lines. Fusion proteins of the hGli2 isoforms with the DNA-binding domain of Gal4 activated transcription when the reporter contained a Gal4-binding site and one copy of the 21-bp sequence, to which CREB binds. This activation was observed only in the presence of Tax. The 21-bp sequence in the reporter was also essential for the activation. These results suggest that simultaneous binding of hGli2 and CREB to the respective sites in the reporter seems to be critical for Tax protein to activate transcription. Consequently, it is probable that the LTR can be regulated by two independent signals through hGli2 and CREB, since the LTR contains the 21-bp and TRE2S sequences in the vicinity.
Figures


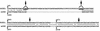


Similar articles
-
Interaction of Gli2 with CREB protein on DNA elements in the long terminal repeat of human T-cell leukemia virus type 1 is responsible for transcriptional activation by tax protein.J Virol. 1999 Apr;73(4):3258-63. doi: 10.1128/JVI.73.4.3258-3263.1999. J Virol. 1999. PMID: 10074179 Free PMC article.
-
A new regulatory element that augments the Tax-dependent enhancer of human T-cell leukemia virus type 1 and cloning of cDNAs encoding its binding proteins.J Virol. 1993 Sep;67(9):5375-82. doi: 10.1128/JVI.67.9.5375-5382.1993. J Virol. 1993. PMID: 8350401 Free PMC article.
-
Protein domains involved in both in vivo and in vitro interactions between human T-cell leukemia virus type I tax and CREB.J Virol. 1995 Jun;69(6):3420-32. doi: 10.1128/JVI.69.6.3420-3432.1995. J Virol. 1995. PMID: 7745688 Free PMC article.
-
Human T cell lymphotropic virus type I genomic expression and impact on intracellular signaling pathways during neurodegenerative disease and leukemia.Front Biosci. 2000 Jan 1;5:D138-68. doi: 10.2741/yao. Front Biosci. 2000. PMID: 10702386 Review.
-
Emerging role of ZBTB7A as an oncogenic driver and transcriptional repressor.Cancer Lett. 2020 Jul 28;483:22-34. doi: 10.1016/j.canlet.2020.04.015. Epub 2020 Apr 26. Cancer Lett. 2020. PMID: 32348807 Review.
Cited by
-
Hedgehog Signaling: Implications in Cancers and Viral Infections.Int J Mol Sci. 2021 Jan 21;22(3):1042. doi: 10.3390/ijms22031042. Int J Mol Sci. 2021. PMID: 33494284 Free PMC article. Review.
-
Loss-of-function mutations in the human GLI2 gene are associated with pituitary anomalies and holoprosencephaly-like features.Proc Natl Acad Sci U S A. 2003 Nov 11;100(23):13424-9. doi: 10.1073/pnas.2235734100. Epub 2003 Oct 27. Proc Natl Acad Sci U S A. 2003. PMID: 14581620 Free PMC article.
-
GLI2 regulates TGF-β1 in human CD4+ T cells: implications in cancer and HIV pathogenesis.PLoS One. 2012;7(7):e40874. doi: 10.1371/journal.pone.0040874. Epub 2012 Jul 31. PLoS One. 2012. PMID: 22859956 Free PMC article.
-
Development of ultra-super sensitive immunohistochemistry and its application to the etiological study of adult T-cell leukemia/lymphoma.Acta Histochem Cytochem. 2012 Apr 26;45(2):83-106. doi: 10.1267/ahc.11034. Epub 2012 Apr 21. Acta Histochem Cytochem. 2012. PMID: 22685351 Free PMC article.
-
Identification of key transcription factors associated with cerebral ischemia‑reperfusion injury based on gene‑set enrichment analysis.Int J Mol Med. 2019 Jun;43(6):2429-2439. doi: 10.3892/ijmm.2019.4159. Epub 2019 Apr 9. Int J Mol Med. 2019. PMID: 31017267 Free PMC article.
References
-
- Chen I S Y, Cann A J, Shah N P, Gaynor R B. Functional relation between HTLV-2 x and adenovirus I1A proteins in transcriptional activation. Science. 1985;230:570–573. - PubMed
-
- Chrivia J C, Kwok R P, Lamb N, Hagiwara M, Montminy M R, Goodman R H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature. 1993;365:855–859. - PubMed
-
- Felber B K, Paskalis H, Kleinman-Ewing D, Wong-Staal F, Pavlakis G N. The pX protein of HTLV-1 is a transcriptional activator of its long terminal repeats. Science. 1985;229:675–679. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

