Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease
- PMID: 9557685
- PMCID: PMC109625
- DOI: 10.1128/JVI.72.5.3980-3990.1998
Natural isolates of simian virus 40 from immunocompromised monkeys display extensive genetic heterogeneity: new implications for polyomavirus disease
Abstract
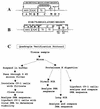
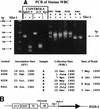
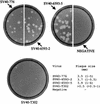
Simian virus 40 (SV40) DNAs in brain tissue and peripheral blood mononuclear cells (PBMCs) of eight simian immunodeficiency virus-infected rhesus monkeys with SV40 brain disease were analyzed. We report the detection, cloning, and identification of five new SV40 strains following a quadruple testing-verification strategy. SV40 genomes with archetypal regulatory regions (containing a duplication within the G/C-rich regulatory region segment and a single 72-bp enhancer element) were recovered from seven animal brains, two tissues of which also contained viral genomes with nonarchetypal regulatory regions (containing a duplication within the G/C-rich regulatory region segment as well as a variable duplication within the enhancer region). In contrast, PBMC DNAs from five of six animals had viral genomes with both regulatory region types. It appeared, based on T-antigen variable-region sequences, that nonarchetypal virus variants arose de novo within each animal. The eighth animal exclusively yielded a new type of SV40 strain (SV40-K661), containing a protoarchetypal regulatory region (lacking a duplication within the G/C-rich segment of the regulatory region and containing one 72-bp element in the enhancer region), from both brain tissue and PBMCs. The presence of SV40 in PBMCs suggests that hematogenous spread of viral infection may occur. An archetypal version of a virus similar to SV40 reference strain 776 (a kidney isolate) was recovered from one brain, substantiating the idea that SV40 is neurotropic as well as kidney-tropic. Indirect evidence suggests that maternal-infant transmission of SV40 may have occurred in one animal. These findings provide new insights for human polyomavirus disease.
Figures



References
-
- Azzi A, De Santis R, Ciappi S, Leoncini F, Sterrantino G, Marino N, Mazzotta F, Laszlo D, Fanci R, Bosi A. Human polyomaviruses DNA detection in peripheral blood leukocytes from immunocompetent and immunocompromised individuals. J Neurovirol. 1996;2:411–416. - PubMed
-
- Bergsagel D J, Finegold M J, Butel J S, Kupsky W J, Garcea R L. DNA sequences similar to those of simian virus 40 in ependymomas and choroid plexus tumors of childhood. N Engl J Med. 1992;326:988–993. - PubMed
-
- Butel J S, Wong C, Medina D. Transformation of mouse mammary epithelial cells by papovavirus SV40. Exp Mol Pathol. 1984;40:79–108. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

