Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target
- PMID: 9557688
- PMCID: PMC109628
- DOI: 10.1128/JVI.72.5.4005-4014.1998
Chromosome structure and human immunodeficiency virus type 1 cDNA integration: centromeric alphoid repeats are a disfavored target
Abstract
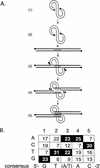
Integration of retroviral cDNA into host chromosomal DNA is an essential and distinctive step in viral replication. Despite considerable study, the host determinants of sites for integration have not been fully clarified. To investigate integration site selection in vivo, we used two approaches. (i) We have analyzed the host sequences flanking 61 human immunodeficiency virus type 1 (HIV-1) integration sites made by experimental infection and compared them to a library of 104 control sequences. (ii) We have also analyzed HIV-1 integration frequencies near several human repeated-sequence DNA families, using a repeat-specific PCR-based assay. At odds with previous reports from smaller-scale studies, we found no strong biases either for or against integration near repetitive sequences such as Alu or LINE-1 elements. We also did not find a clear bias for integration in transcription units as proposed previously, although transcription units were found somewhat more frequently near integration sites than near controls. However, we did find that centromeric alphoid repeats were selectively absent at integration sites. The repeat-specific PCR-based assay also indicated that alphoid repeats were disfavored for integration in vivo but not as naked DNA in vitro. Evidently the distinctive DNA organization at centromeres disfavors cDNA integration. We also found a weak consensus sequence for host DNA at integration sites, and assays of integration in vitro indicated that this sequence is favored as naked DNA, revealing in addition an influence of target primary sequence.
Figures
References
-
- Boeke J D. Transposable elements in Saccharomyces cerevisiae. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 335–374.
-
- Bor Y-C, Miller M, Bushman F, Orgel L. Target sequence preferences of HIV-1 integration complexes in vitro. Virology. 1996;222:238–242. - PubMed
-
- Brown P O, Bowerman B, Varmus H E, Bishop J M. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

