A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release
- PMID: 9557699
- PMCID: PMC109639
- DOI: 10.1128/JVI.72.5.4095-4103.1998
A proline-rich motif (PPPY) in the Gag polyprotein of Mason-Pfizer monkey virus plays a maturation-independent role in virion release
Abstract
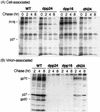
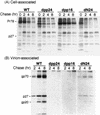
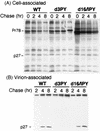
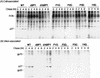
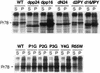
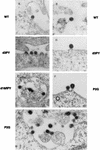
Virus assembly represents one of the last steps in the retrovirus life cycle. During this process, Gag polyproteins assemble at specific sites within the cell to form viral capsids and induce membrane extrusion (viral budding) either as assembly progresses (type C virus) or following formation of a complete capsid (type B and type D viruses). Finally, the membrane must undergo a fusion event to pinch off the particle in order to release a complete enveloped virion. Structural elements within the MA region of the Gag polyprotein define the route taken to the plasma membrane and direct the process of virus budding. Results presented here suggest that a distinct region of Gag is necessary for virus release. The pp24 and pp16 proteins of the type D retrovirus Mason-Pfizer monkey virus (M-PMV) are phosphoproteins that are encoded in the gag gene of the virus. The pp16 protein is a C-terminally located cleavage product of pp24 and contains a proline-rich motif (PPPY) that is conserved among the Gag proteins of a wide variety of retroviruses. By performing a functional analysis of this coding region with deletion mutants, we have shown that the pp16 protein is dispensable for capsid assembly but essential for virion release. Moreover, additional experiments indicated that the virus release function of pp16 was abolished by the deletion of only the PPPY motif and could be restored when this motif alone was reinserted into a Gag polyprotein lacking the entire pp16 domain. Single-amino-acid substitutions for any of the residues within this motif confer a similar virion release-defective phenotype. It is unlikely that the function of the proline-rich motif is simply to inhibit premature activation of protease, since the PPPY deletion blocked virion release in the context of a protease-defective provirus. These results demonstrate that in type D retroviruses a PPPY motif plays a key role in a late stage of virus budding that is independent of and occurs prior to virion maturation.
Figures

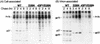
References
-
- Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. - PubMed
-
- Bowles N, Bonnet D, Mulhauser F, Spahr P-F. Site-directed mutagenesis of the P2 region of the Rous sarcoma virus gag gene: effects on Gag polyprotein processing. Virology. 1994;203:20–28. - PubMed
-
- Bradac J, Hunter E. Polypeptides of Mason-Pfizer monkey virus. I. Synthesis and processing of the gag-gene products. Virology. 1984;138:260–275. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

