Use of a rapid throughput in vivo screen to investigate inhibitors of eukaryotic topoisomerase II enzymes
- PMID: 9559802
- PMCID: PMC105561
- DOI: 10.1128/AAC.42.4.889
Use of a rapid throughput in vivo screen to investigate inhibitors of eukaryotic topoisomerase II enzymes
Abstract
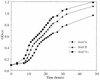
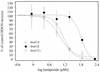
Topoisomerase II catalyzes the passage of one DNA helix through another via a transient double-stranded break. The essential nature of this enzyme in cell proliferation and its mechanism of action make it an ideal target for cytotoxic agents. Saccharomyces cerevisiae topoisomerase II has been frequently used as a model for testing potential inhibitors of eukaryotic topoisomerase II as antitumor agents. The standard in vivo method of estimating the sensitivity of S. cerevisiae to the antitopoisomerase drugs is via inhibition or kill curves which rely on viable-cell counts and is labor intensive. We present an alternative to this, a high-throughput in vivo screen. This method makes use of a drug-permeable S. cerevisiae strain lacking endogenous topoisomerase II, which is modified to express either human topoisomerase IIalpha or IIbeta or S. cerevisiae topoisomerase II carried on plasmids. Each modified strain expresses a full-length topoisomerase II enzyme, as opposed to the more commonly used temperature-sensitive S. cerevisiae mutant expressing yeast or yeast/human hybrid enzymes. A comparison of this new method with a plating-and-counting method gave similar drug sensitivity results, with increased accuracy and reduced manual input for the new method. The information generated has highlighted the sensitivities of different topoisomerase II enzymes and isoenzymes to several different classes of topoisomerase II inhibitor.
Figures

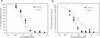

References
-
- Berger J M, Gamblin S J, Harrison S C, Wang J C. Structure and mechanism of DNA topoisomerase-II. Nature. 1996;379:225–232. - PubMed
-
- Chen A Y, Liu L F. DNA topoisomerases: essential enzymes and lethal targets. Annu Rev Pharmacol Toxicol. 1994;34:191–218. - PubMed
-
- Cornarotti M, Tinelli S, Willmore E, Zunino F, Fisher L M, Austin C A, Capranico G. Drug sensitivity and sequence specificity of human recombinant DNA topoisomerases II alpha (p170) and II beta (p180) Mol Pharmacol. 1996;50:1463–1471. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

