A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase
- PMID: 9560209
- PMCID: PMC20194
- DOI: 10.1073/pnas.95.9.4953
A stationary phase protein in Escherichia coli with binding activity to the major sigma subunit of RNA polymerase
Abstract
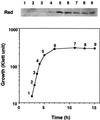
Switching of the transcription pattern in Escherichia coli during the growth transition from exponential to stationary phase is accompanied by the replacement of the RNA polymerase-associated sigma70 subunit (sigmaD) with sigma38 (sigmaS). A fraction of the sigma70 subunit in stationary phase cell extracts was found to exist as a complex with a novel protein, designated Rsd (Regulator of sigma D). The intracellular level of Rsd starts to increase during the transition from growing to stationary phase. The rsd gene was identified at 90 min on the E. coli chromosome. Overexpressed and purified Rsd protein formed complexes in vitro with sigma70 but not with other sigma subunits, sigmaN, sigmaS, sigmaH, sigmaF, and sigmaE. Analysis of proteolytic fragments of sigma70 indicated that Rsd binds at or downstream of region 4, the promoter -35 recognition domain. The isolated Rsd inhibited transcription in vitro to various extents depending on the promoters used. We propose that Rsd is a stationary phase E. coli protein with regulatory activity of the sigma70 function.
Figures




References
-
- Helmann J D, Chamberlin M J. Annu Rev Biochem. 1988;57:839–872. - PubMed
-
- Gross C A, Lonetto M, Losick R. In: Transcription Regulation. McKnight S L, Yamamoto K R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1992. pp. 129–176.
-
- Ishihama A. In: Mechanisms of Transcription. Eckstein F, Lilley D M J, editors. Berlin: Springer; 1997. pp. 53–70.
-
- Ishihama A, Taketo M, Saitoh T, Fukuda R. In: RNA Polymerase. Chamberlin M, Losick R, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1976. pp. 475–502.
-
- Ishihama A. In: Control of Cell Growth and Division. Ishihama A, Yoshikawa H, editors. Heidelberg: Springer; 1991. pp. 121–140.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

