Are protein folds atypical?
- PMID: 9560215
- PMCID: PMC20200
- DOI: 10.1073/pnas.95.9.4987
Are protein folds atypical?
Abstract
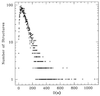
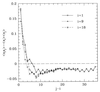
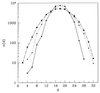
Protein structures are a very special class among all possible structures. It has been suggested that a "designability principle" plays a crucial role in nature's selection of protein sequences and structures. Here, we provide a theoretical base for such a selection principle, using a simple model of protein folding based on hydrophobic interactions. A structure is reduced to a string of 0s and 1s, which represent the surface and core sites, respectively, as the backbone is traced. Each structure is therefore associated with one point in a high dimensional space. Sequences are represented by strings of their hydrophobicities and thus can be mapped into the same space. A sequence that lies closer to a particular structure in this space than to any other structures will have that structure as its ground state. Atypical structures, namely those far away from other structures in the high dimensional space, have more sequences that fold into them and are thermodynamically more stable. We argue that the most common folds of proteins are the most atypical in the space of possible structures.
Figures


Similar articles
-
The designability of protein structures.J Mol Graph Model. 2001;19(1):157-67. doi: 10.1016/s1093-3263(00)00137-6. J Mol Graph Model. 2001. PMID: 11381527
-
Exploration of the relationship between topology and designability of conformations.J Chem Phys. 2011 Jun 21;134(23):235101. doi: 10.1063/1.3596947. J Chem Phys. 2011. PMID: 21702580 Free PMC article.
-
Designable structures are easy to unfold.Phys Rev E Stat Nonlin Soft Matter Phys. 2006 Oct;74(4 Pt 1):042902. doi: 10.1103/PhysRevE.74.042902. Epub 2006 Oct 9. Phys Rev E Stat Nonlin Soft Matter Phys. 2006. PMID: 17155116
-
The family feud: do proteins with similar structures fold via the same pathway?Curr Opin Struct Biol. 2005 Feb;15(1):42-9. doi: 10.1016/j.sbi.2005.01.011. Curr Opin Struct Biol. 2005. PMID: 15718132 Review.
-
Inter-residue interactions in protein folding and stability.Prog Biophys Mol Biol. 2004 Oct;86(2):235-77. doi: 10.1016/j.pbiomolbio.2003.09.003. Prog Biophys Mol Biol. 2004. PMID: 15288760 Review.
Cited by
-
Thoroughly sampling sequence space: large-scale protein design of structural ensembles.Protein Sci. 2002 Dec;11(12):2804-13. doi: 10.1110/ps.0203902. Protein Sci. 2002. PMID: 12441379 Free PMC article.
-
Folding protein models with a simple hydrophobic energy function: the fundamental importance of monomer inside/outside segregation.Proc Natl Acad Sci U S A. 1999 Oct 26;96(22):12482-7. doi: 10.1073/pnas.96.22.12482. Proc Natl Acad Sci U S A. 1999. PMID: 10535948 Free PMC article.
-
Coarse-grained sequences for protein folding and design.Proc Natl Acad Sci U S A. 2003 Sep 16;100(19):10712-7. doi: 10.1073/pnas.1931882100. Epub 2003 Sep 8. Proc Natl Acad Sci U S A. 2003. PMID: 12963815 Free PMC article.
-
Designability of alpha-helical proteins.Proc Natl Acad Sci U S A. 2002 Aug 20;99(17):11163-8. doi: 10.1073/pnas.162105999. Epub 2002 Aug 12. Proc Natl Acad Sci U S A. 2002. PMID: 12177419 Free PMC article.
-
Comparative modeling and protein-like features of hydrophobic-polar models on a two-dimensional lattice.Proteins. 2012 Jun;80(6):1683-93. doi: 10.1002/prot.24067. Epub 2012 Apr 13. Proteins. 2012. PMID: 22411636 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

