Targeted disruption of p70(s6k) defines its role in protein synthesis and rapamycin sensitivity
- PMID: 9560223
- PMCID: PMC20208
- DOI: 10.1073/pnas.95.9.5033
Targeted disruption of p70(s6k) defines its role in protein synthesis and rapamycin sensitivity
Abstract
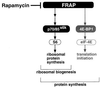
Here, we disrupted the p70 S6 kinase (p70(s6k)) gene in murine embryonic stem cells to determine the role of this kinase in cell growth, protein synthesis, and rapamycin sensitivity. p70(s6k-/-) cells proliferated at a slower rate than parental cells, suggesting that p70(s6k) has a positive influence on cell proliferation but is not essential. In addition, rapamycin inhibited proliferation of p70(s6k-/-) cells, indicating that other events inhibited by the drug, independent of p70(s6k), also are important for both cell proliferation and the action of rapamycin. In p70(s6k-/-) cells, which exhibited no ribosomal S6 phosphorylation, translation of mRNA encoding ribosomal proteins was not increased by serum nor specifically inhibited by rapamycin. In contrast, rapamycin inhibited phosphorylation of initiation factor 4E-binding protein 1 (4E-BP1), general mRNA translation, and overall protein synthesis in p70(s6k-/-) cells, indicating that these events proceed independently of p70(s6k) activity. This study localizes the function of p70(s6k) to ribosomal biogenesis by regulating ribosomal protein synthesis at the level of mRNA translation.
Figures




References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

