Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans
- PMID: 9560244
- PMCID: PMC20229
- DOI: 10.1073/pnas.95.9.5150
Monosomy of a specific chromosome determines L-sorbose utilization: a novel regulatory mechanism in Candida albicans
Abstract
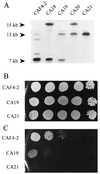
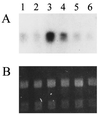
We report the identification of the gene, SOU1, required for L-sorbose assimilation in Candida albicans. The level of the expression of SOU1 is determined by the copy number of chromosome III (also denoted chromosome 5), such that monosomic strains assimilate L-sorbose, whereas disomic strains do not, in spite of the fact that SOU1 is not on this chromosome. We suggest that C. albicans contains a resource of potentially beneficial genes that are activated by changes in chromosome number, and that this elaborate mechanism regulates the utilization of food supplies and possibly other important functions, thus representing a novel general means for regulating gene expression in microbes.
Figures
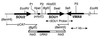


References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

