Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors
- PMID: 9560288
- PMCID: PMC20273
- DOI: 10.1073/pnas.95.9.5401
Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors
Erratum in
- Proc Natl Acad Sci U S A 1998 Jul 21;95(15):9060
Abstract
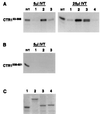
In Arabidopsis thaliana, signal transduction of the hormone ethylene involves at least two receptors, ETR1 and ERS, both of which are members of the two-component histidine protein kinase family that is prevalent in prokaryotes. The pathway also contains a negative regulator of ethylene responses, CTR1, which closely resembles members of the Raf protein kinase family. CTR1 is thought to act at or downstream of ETR1 and ERS based on double mutant analysis; however, the signaling mechanisms leading from ethylene perception to the regulation of CTR1 are unknown. By using the yeast two-hybrid assay, we detected a specific interaction between the CTR1 amino-terminal domain and the predicted histidine kinase domain of ETR1 and ERS. We subsequently verified these interactions by using an in vitro protein association assay(s). In addition, we determined that the amino-terminal domain of CTR1 can associate with the predicted receiver domain of ETR1 in vitro. Based on deletion analysis, the portion of CTR1 that interacts with ETR1 roughly aligns with the regulatory region of Raf kinases. These physical associations support the genetic evidence that CTR1 acts in the pathway of ETR1 and ERS and suggest that these interactions could be involved in the regulation of CTR1 activity.
Figures



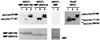
References
-
- Abeles F B, Morgan P W, Saltveit M E., Jr . Ethylene in Plant Biology. 2nd Ed. New York: Academic; 1992.
-
- Chang C, Kwok S F, Bleecker A B, Meyerowitz E M. Science. 1993;262:539–544. - PubMed
-
- Hua J, Chang C, Sun Q, Meyerowitz E M. Science. 1995;269:1712–1714. - PubMed
-
- Kieber J J, Rothenberg M, Roman G, Feldmann K A, Ecker J R. Cell. 1993;72:427–441. - PubMed
-
- Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker J R. Cell. 1997;89:1133–1144. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

