Stat3 activation by Src induces specific gene regulation and is required for cell transformation
- PMID: 9566874
- PMCID: PMC110634
- DOI: 10.1128/MCB.18.5.2545
Stat3 activation by Src induces specific gene regulation and is required for cell transformation
Abstract
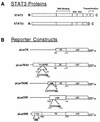
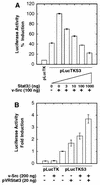
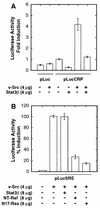
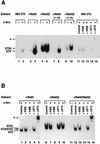
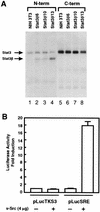
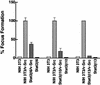
While signal transducers and activators of transcription (STATs) were originally discovered as intracellular effectors of normal signaling by cytokines, increasing evidence also points to a role for STAT transcription factors in oncogenesis. Previous studies have demonstrated that one STAT family member, Stat3, possesses constitutively elevated tyrosine phosphorylation and DNA-binding activity in fibroblasts stably transformed by the Src oncoprotein. To determine if this Stat3 activation by Src could induce Stat3-mediated gene expression, luciferase reporter constructs based on synthetic and authentic promoters were transfected into NIH 3T3 cells. Activation of endogenous cellular Stat3 by the Src oncoprotein induced gene expression through a Stat3-specific binding element (TTCCCGAA) of the C-reactive protein gene promoter. A naturally occurring splice variant of human Stat3 protein, Stat3beta, with a deletion in the C-terminal transactivation domain abolished this gene induction in a dominant negative manner. Expression of Stat3beta did not have any effect on a reporter construct based on the c-fos serum response element, which is not dependent on Stat3 signaling, indicating that Stat3beta does not nonspecifically inhibit other signaling pathways or Src function. Transfection of vectors expressing Stat3beta together with Src blocked cell transformation by Src as measured in a quantitative focus formation assay using NIH 3T3 cells. By contrast, Stat3beta had a much less pronounced effect on focus formation induced by the Ras oncoprotein, which does not activate Stat3 signaling. In addition, three independent clones of NIH 3T3 cells stably overexpressing Stat3beta were generated and characterized, demonstrating that Stat3beta overexpression does not have a toxic effect on cell viability. These Stat3beta-overexpressing clones were shown to be deficient in Stat3-mediated signaling and refractory to Src-induced cell transformation. We conclude that Stat3 activation by the Src oncoprotein leads to specific gene regulation and that Stat3 is one of the critical signaling pathways involved in Src oncogenesis. Our findings provide evidence that oncogenesis-associated activation of Stat3 signaling is part of the process of malignant transformation.
Figures



References
-
- Akira S, Nishio Y, Inoue M, Wang X-J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Molecular cloning of APRF, a novel IFN-stimulated gene factor 3 p91-related transcription factor involved in the gp130-mediated signaling pathway. Cell. 1994;77:63–71. - PubMed
-
- Bowman, T., and R. Jove. Unpublished data.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
