The second catalytic domain of protein tyrosine phosphatase delta (PTP delta) binds to and inhibits the first catalytic domain of PTP sigma
- PMID: 9566880
- PMCID: PMC110640
- DOI: 10.1128/MCB.18.5.2608
The second catalytic domain of protein tyrosine phosphatase delta (PTP delta) binds to and inhibits the first catalytic domain of PTP sigma
Abstract
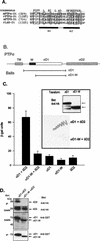
The LAR family protein tyrosine phosphatases (PTPs), including LAR, PTP delta, and PTP sigma, are transmembrane proteins composed of a cell adhesion molecule-like ectodomain and two cytoplasmic catalytic domains: active D1 and inactive D2. We performed a yeast two-hybrid screen with the first catalytic domain of PTP sigma (PTP sigma-D1) as bait to identify interacting regulatory proteins. Using this screen, we identified the second catalytic domain of PTP delta (PTP delta-D2) as an interactor of PTP sigma-D1. Both yeast two-hybrid binding assays and coprecipitation from mammalian cells revealed strong binding between PTP sigma-D1 and PTP delta-D2, an association which required the presence of the wedge sequence in PTP sigma-D1, a sequence recently shown to mediate D1-D1 homodimerization in the phosphatase RPTP alpha. This interaction was not reciprocal, as PTP delta-D1 did not bind PTP sigma-D2. Addition of a glutathione S-transferase (GST)-PTP delta-D2 fusion protein (but not GST alone) to GST-PTP sigma-D1 led to approximately 50% inhibition of the catalytic activity of PTP sigma-D1, as determined by an in vitro phosphatase assay against p-nitrophenylphosphate. A similar inhibition of PTP sigma-D1 activity was obtained with coimmunoprecipitated PTP delta-D2. Interestingly, the second catalytic domains of LAR (LAR-D2) and PTP sigma (PTP sigma-D2), very similar in sequence to PTP delta-D2, bound poorly to PTP sigma-D1. PTP delta-D1 and LAR-D1 were also able to bind PTP delta-D2, but more weakly than PTP sigma-D1, with a binding hierarchy of PTP sigma-D1 >> PTP delta-D1 > LAR-D1. These results suggest that association between PTP sigma-D1 and PTP delta-D2, possibly via receptor heterodimerization, provides a negative regulatory function and that the second catalytic domains of this and likely other receptor PTPs, which are often inactive, may function instead to regulate the activity of the first catalytic domains.
Figures






References
-
- Barnea G, Silvennoinen O, Shaanan B, Honegger A M, Canoll P D, D’Eustachio P, Morse B, Levy J B, LaForgia S, Huebner K, Musacchio J M, Sap J, Schlessinger J. Identification of a carbonic anhydrase-like domain in the extracellular region of RPTPγ defines a new subfamily of receptor tyrosine phosphatases. Mol Cell Biol. 1993;13:1497–1506. - PMC - PubMed
-
- Bilwes A M, den Hertog J, Hunter T, Noel J P. Structural basis for inhibition of receptor protein-tyrosine-α phosphatase by dimerization. Nature. 1996;382:555–559. - PubMed
-
- Brady-Kalnay S M, Tonks N K. Identification of the homophilic binding site of the receptor protein tyrosine phosphatase PTPμ. J Biol Chem. 1994;269:28472–28477. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
