Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2
- PMID: 9566882
- PMCID: PMC110642
- DOI: 10.1128/MCB.18.5.2629
Cooperative Pho2-Pho4 interactions at the PHO5 promoter are critical for binding of Pho4 to UASp1 and for efficient transactivation by Pho4 at UASp2
Abstract
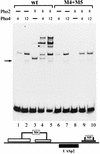
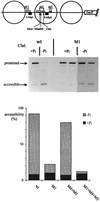
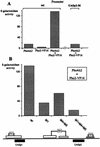
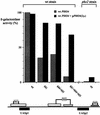
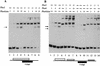
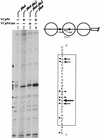
The activation of the PHO5 gene in Saccharomyces cerevisiae in response to phosphate starvation critically depends on two transcriptional activators, the basic helix-loop-helix protein Pho4 and the homeodomain protein Pho2. Pho4 acts through two essential binding sites corresponding to the regulatory elements UASp1 and UASp2. Mutation of either of them results in a 10-fold decrease in promoter activity, and mutation of both sites renders the promoter totally uninducible. The role of Pho4 appears relatively straightforward, but the mechanism of action of Pho2 had remained elusive. By in vitro footprinting, we have recently mapped multiple Pho2 binding sites adjacent to the Pho4 sites, and by mutating them individually or in combination, we now show that each of them contributes to PHO5 promoter activity. Their function is not only to recruit Pho2 to the promoter but to allow cooperative binding of Pho4 together with Pho2. Cooperativity requires DNA binding of Pho2 to its target sites and Pho2-Pho4 interactions. A Pho4 derivative lacking the Pho2 interaction domain is unable to activate the promoter, but testing of UASp1 and UASp2 individually in a minimal CYC1 promoter reveals a striking difference between the two UAS elements. UASp1 is fully inactive, presumably because the Pho4 derivative is not recruited to its binding site. In contrast, UASp2 activates strongly in a Pho2-independent manner. From in vivo footprinting experiments and activity measurements with a promoter variant containing two UASp2 elements, we conclude that at UASp2, Pho2 is mainly required for the ability of Pho4 to transactivate.
Figures




References
-
- Arndt K T, Styles C, Fink G R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987;237:874–880. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
