Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis
- PMID: 9566888
- PMCID: PMC110648
- DOI: 10.1128/MCB.18.5.2688
Ski6p is a homolog of RNA-processing enzymes that affects translation of non-poly(A) mRNAs and 60S ribosomal subunit biogenesis
Abstract
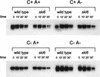
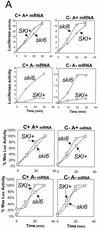
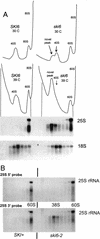
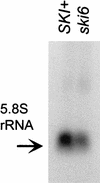
We mapped and cloned SKI6 of Saccharomyces cerevisiae, a gene that represses the copy number of the L-A double-stranded RNA virus, and found that it encodes an essential 246-residue protein with homology to a tRNA-processing enzyme, RNase PH. The ski6-2 mutant expressed electroporated non-poly(A) luciferase mRNAs 8- to 10-fold better than did the isogenic wild type. No effect of ski6-2 on expression of uncapped or normal mRNAs was found. Kinetics of luciferase synthesis and direct measurement of radiolabeled electroporated mRNA indicate that the primary effect of Ski6p was on efficiency of translation rather than on mRNA stability. Both ski6 and ski2 mutants show hypersensitivity to hygromycin, suggesting functional alteration of the translation apparatus. The ski6-2 mutant has normal amounts of 40S and 60S ribosomal subunits but accumulates a 38S particle containing 5'-truncated 25S rRNA but no 5.8S rRNA, apparently an incomplete or degraded 60S subunit. This suggests an abnormality in 60S subunit assembly. The ski6-2 mutation suppresses the poor expression of the poly(A)- viral mRNA in a strain deficient in the 60S ribosomal protein L4. Thus, a ski6 mutation bypasses the requirement of the poly(A) tail for translation, allowing better translation of non-poly(A) mRNA, including the L-A virus mRNA which lacks poly(A). We speculate that the derepressed translation of non-poly(A) mRNAs is due to abnormal (but full-size) 60S subunits.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
