Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA
- PMID: 9566912
- PMCID: PMC110672
- DOI: 10.1128/MCB.18.5.2932
Selenium deficiency reduces the abundance of mRNA for Se-dependent glutathione peroxidase 1 by a UGA-dependent mechanism likely to be nonsense codon-mediated decay of cytoplasmic mRNA
Abstract
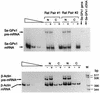
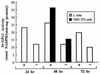
The mammalian mRNA for selenium-dependent glutathione peroxidase 1 (Se-GPx1) contains a UGA codon that is recognized as a codon for the nonstandard amino acid selenocysteine (Sec). Inadequate concentrations of selenium (Se) result in a decrease in Se-GPx1 mRNA abundance by an uncharacterized mechanism that may be dependent on translation, independent of translation, or both. In this study, we have begun to elucidate this mechanism. We demonstrate using hepatocytes from rats fed either a Se-supplemented or Se-deficient diet for 9 to 13 weeks that Se deprivation results in an approximately 50-fold reduction in Se-GPx1 activity and an approximately 20-fold reduction in Se-GPx1 mRNA abundance. Reverse transcription-PCR analyses of nuclear and cytoplasmic fractions revealed that Se deprivation has no effect on the levels of either nuclear pre-mRNA or nuclear mRNA but reduces the level of cytoplasmic mRNA. The regulation of Se-GPx1 gene expression by Se was recapitulated in transient transfections of NIH 3T3 cells, and experiments were extended to examine the consequences of converting the Sec codon (TGA) to either a termination codon (TAA) or a cysteine codon (TGC). Regardless of the type of codon, an alteration in the Se concentration was of no consequence to the ratio of nuclear Se-GPx1 mRNA to nuclear Se-GPx1 pre-mRNA. The ratio of cytoplasmic Se-GPx1 mRNA to nuclear Se-GPx1 mRNA from the wild-type (TGA-containing) allele was reduced twofold when cells were deprived of Se for 48 h after transfection, which has been shown to be the extent of the reduction for the endogenous Se-GPx1 mRNA of cultured cells incubated as long as 20 days in Se-deficient medium. In contrast to the TGA allele, Se had no effect on expression of either the TAA allele or the TGC allele. Under Se-deficient conditions, the TAA and TGC alleles generated, respectively, 1.7-fold-less and 3-fold-more cytoplasmic Se-GPx1 mRNA relative to the amount of nuclear Se-GPx1 mRNA than the TGA allele. These results indicate that (i) under conditions of Se deprivation, the Sec codon reduces the abundance of cytoplasmic Se-GPx1 mRNA by a translation-dependent mechanism and (ii) there is no additional mechanism by which Se regulates Se-GPx1 mRNA production. These data suggest that the inefficient incorporation of Sec at the UGA codon during mRNA translation augments the nonsense-codon-mediated decay of cytoplasmic Se-GPx1 mRNA.
Figures


Similar articles
-
Nonsense-mediated decay of mRNA for the selenoprotein phospholipid hydroperoxide glutathione peroxidase is detectable in cultured cells but masked or inhibited in rat tissues.Mol Biol Cell. 2001 Apr;12(4):1009-17. doi: 10.1091/mbc.12.4.1009. Mol Biol Cell. 2001. PMID: 11294903 Free PMC article.
-
Cis-acting elements are required for selenium regulation of glutathione peroxidase-1 mRNA levels.RNA. 1998 Jul;4(7):816-27. doi: 10.1017/s1355838298971990. RNA. 1998. PMID: 9671054 Free PMC article.
-
Nonsense-mediated decay of glutathione peroxidase 1 mRNA in the cytoplasm depends on intron position.EMBO J. 2000 Sep 1;19(17):4734-44. doi: 10.1093/emboj/19.17.4734. EMBO J. 2000. PMID: 10970865 Free PMC article.
-
Evidence that selenium deficiency results in the cytoplasmic decay of GPx1 mRNA dependent on pre-mRNA splicing proteins bound to the mRNA exon-exon junction.Biofactors. 2001;14(1-4):37-42. doi: 10.1002/biof.5520140106. Biofactors. 2001. PMID: 11568438 Review.
-
The beginning of GPX2 and 30 years later.Free Radic Biol Med. 2022 Aug 1;188:419-433. doi: 10.1016/j.freeradbiomed.2022.06.232. Epub 2022 Jul 5. Free Radic Biol Med. 2022. PMID: 35803440 Free PMC article. Review.
Cited by
-
Post-transcriptional defects of antioxidant selenoenzymes cause oxidative stress under methylmercury exposure.J Biol Chem. 2011 Feb 25;286(8):6641-9. doi: 10.1074/jbc.M110.168872. Epub 2010 Nov 24. J Biol Chem. 2011. PMID: 21106535 Free PMC article.
-
The multiple lives of NMD factors: balancing roles in gene and genome regulation.Nat Rev Genet. 2008 Sep;9(9):699-712. doi: 10.1038/nrg2402. Nat Rev Genet. 2008. PMID: 18679436 Free PMC article. Review.
-
A system for coordinated analysis of translational readthrough and nonsense-mediated mRNA decay.PLoS One. 2017 Mar 21;12(3):e0173980. doi: 10.1371/journal.pone.0173980. eCollection 2017. PLoS One. 2017. PMID: 28323884 Free PMC article.
-
Selenium regulation of transcript abundance and translational efficiency of glutathione peroxidase-1 and -4 in rat liver.Biochem J. 2001 Aug 1;357(Pt 3):851-8. doi: 10.1042/0264-6021:3570851. Biochem J. 2001. PMID: 11463357 Free PMC article.
-
Translational regulation of GPx-1 and GPx-4 by the mTOR pathway.PLoS One. 2014 Apr 1;9(4):e93472. doi: 10.1371/journal.pone.0093472. eCollection 2014. PLoS One. 2014. PMID: 24691473 Free PMC article.
References
-
- Baker R D, Baker S S, LaRosa K, Whitney C, Newburger P E. Selenium regulation of glutathione peroxidase in human hepatoma cell line Hep3B. Arch Biochem Biophys. 1993;304:53–57. - PubMed
-
- Behne D, Wolters W. Distribution of selenium and glutathione peroxidase in the rat. J Nutr. 1983;113:456–461. - PubMed
-
- Behne D, Hilmert H, Scheid S, Gessner H, Elger W. Evidence for specific selenium target tissues and new biologically important selenoproteins. Biochim Biophys Acta. 1988;966:12–21. - PubMed
-
- Bermano G, Arthur J R, Hesketh J E. Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mRNA stability by selenium supply. FEBS Lett. 1996;387:157–160. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
