Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1
- PMID: 9566914
- PMCID: PMC110674
- DOI: 10.1128/MCB.18.5.2949
Nuclear receptor DAX-1 recruits nuclear receptor corepressor N-CoR to steroidogenic factor 1
Abstract
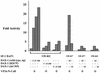
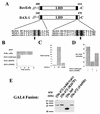
The orphan nuclear receptor steroidogenic factor 1 (SF-1) is a critical developmental regulator in the urogenital ridge, because mice targeted for disruption of the SF-1 gene lack adrenal glands and gonads. SF-1 was recently shown to interact with DAX-1, another orphan receptor whose tissue distribution overlaps that of SF-1. Naturally occurring loss-of-function mutations of the DAX-1 gene cause the human disorder X-linked adrenal hypoplasia congenita (AHC), which resembles the phenotype of SF-1-deficient mice. Paradoxically, however, DAX-1 represses the transcriptional activity of SF-1, and AHC mutants of DAX-1 lose repression function. To further investigate these findings, we characterized the interaction between SF-1 and DAX-1 and found that their interaction indeed occurs through a repressive domain within the carboxy terminus of SF-1. Furthermore, we demonstrate that DAX-1 recruits the nuclear receptor corepressor N-CoR to SF-1, whereas naturally occurring AHC mutations of DAX-1 permit the SF-1-DAX-1 interaction, but markedly diminish corepressor recruitment. Finally, the interaction between DAX-1 and N-CoR shares similarities with that of the nuclear receptor RevErb and N-CoR, because the related corepressor SMRT was not efficiently recruited by DAX-1. Therefore, DAX-1 can serve as an adapter molecule that recruits nuclear receptor corepressors to DNA-bound nuclear receptors like SF-1, thereby extending the range of corepressor action.
Figures





References
-
- Alland L, Muhle R, Hou J, Potes J J, Chin L, Schrieber-Agus N, DePinho R. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. - PubMed
-
- Bae D S, Schaefer M L, Partan B W, Muglia L. Characterization of the mouse DAX-1 gene reveals evolutionary conservation of a unique amino-terminal motif and widespread expression in mouse tissue. Endocrinology. 1996;137:3921–3927. - PubMed
-
- Bakke M, Lund J. Mutually exclusive interactions of two nuclear orphan receptors determine activity of a cAMP-responsive sequence in the bovine CYP17 gene. Mol Endocrinol. 1995;9:327–339. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
