Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1alpha knockout mouse
- PMID: 9566924
- PMCID: PMC110684
- DOI: 10.1128/MCB.18.5.3059
Laron dwarfism and non-insulin-dependent diabetes mellitus in the Hnf-1alpha knockout mouse
Abstract
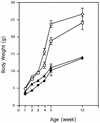
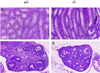
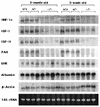
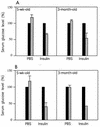
Mice deficient in hepatocyte nuclear factor 1 alpha (HNF-1alpha) were produced by use of the Cre-loxP recombination system. HNF-1alpha-null mice are viable but sterile and exhibit a phenotype reminiscent of both Laron-type dwarfism and non-insulin-dependent diabetes mellitus (NIDDM). In contrast to an earlier HNF-1alpha-null mouse line that had been produced by use of standard gene disruption methodology (M. Pontoglio, J. Barra, M. Hadchouel, A. Doyen, C. Kress, J. P. Bach, C. Babinet, and M. Yaniv, Cell 84:575-585, 1996), these mice exhibited no increased mortality and only minimal renal dysfunction during the first 6 months of development. Both dwarfism and NIDDM are most likely due to the loss of expression of insulin-like growth factor I (IGF-I) and lower levels of insulin, resulting in stunted growth and elevated serum glucose levels, respectively. These results confirm the functional significance of the HNF-1alpha regulatory elements that had previously been shown to reside in the promoter regions of both the IGF-I and the insulin genes.
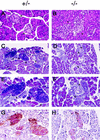
Figures



References
-
- Aden D P, Fogel A, Plotkin S, Damjanov I, Knowles B B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615–616. - PubMed
-
- Artelt P, Grannemann R, Stocking C, Friel J, Bartsch J, Hauser H. The prokaryotic neomycin-resistance-encoding gene acts as a transcriptional silencer in eukaryotic cells. Gene. 1991;99:249–254. - PubMed
-
- Baker J, Hardy M P, Zhou J, Bondy C, Lupu F, Bellve A R, Efstratiadis A. Effects of an Igf1 gene null mutation on mouse reproduction. Mol Endocrinol. 1996;10:903–918. - PubMed
-
- Baumhueter S, Mendel D B, Conley P B, Kuo C J, Turk C, Graves M K, Edwards C A, Courtois G, Crabtree G R. HNF-1 shares three sequence motifs with the POU domain proteins and is identical to LF-B1 and APF. Genes Dev. 1990;4:372–379. - PubMed
-
- Blumenfeld M, Maury M, Chouard T, Yaniv M, Condamine H. Hepatic nuclear factor 1 (HNF 1) shows a wider distribution than products of its known target genes in developing mouse. Development. 1991;113:589–599. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous
