Effect of carbohydrate position on lysosomal transport of procathepsin L
- PMID: 9571245
- PMCID: PMC25336
- DOI: 10.1091/mbc.9.5.1135
Effect of carbohydrate position on lysosomal transport of procathepsin L
Abstract
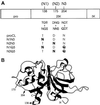
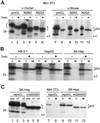
To study the role of carbohydrate in lysosomal protein transport, we engineered two novel glycosylation signals (Asn-X-Ser/Thr) into the cDNA of human procathepsin L, a lysosomal acid protease. We constructed six mutant cDNAs encoding glycosylation signals at mutant sites Asn-138, Asn-175, or both sites together, in the presence or absence of the wild-type Asn-204 site. We stably transfected wild-type and mutant cDNAs into NIH3T3 mouse fibroblasts and then used species-specific antibodies to determine the glycosylation status, phosphorylation, localization, and transport kinetics of recombinant human procathepsin L containing one, two, or three glycosylation sites. Both novel glycosylation sites were capable of being glycosylated, although Asn-175 was utilized only 30-50% of the time. Like the wild-type glycosylation at Asn-204, carbohydrates at Asn-138 and Asn-175 were completely sensitive to endoglycosidase H, and they were phosphorylated. Mutant proteins containing two carbohydrates were capable of being delivered to lysosomes, but there was not a consistent relationship between the efficiency of lysosomal delivery and carbohydrate content of the protein. Pulse-chase labeling revealed a unique biosynthetic pattern for proteins carrying the Asn-175 glycosylation sequence. Whereas wild-type procathepsin L and mutants bearing carbohydrate at Asn-138 appeared in lysosomes by about 60 min, proteins with carbohydrate at Asn-175 were processed to a lysosome-like polypeptide within 15 min. Temperature shift, brefeldin A, and NH4Cl experiments suggested that the rapid processing did not occur in the endoplasmic reticulum and that Asn-175 mutants could interact with the mannose 6-phosphate receptor. Taken together, our results are consistent with the interpretation that Asn-175 carbohydrate confers rapid transport to lysosomes. We may have identified a recognition domain in procathepsin L that is important for its interactions with the cellular transport machinery.
Figures





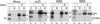
Similar articles
-
Mouse procathepsin L lacking a functional glycosylation site is properly folded, stable, and secreted by NIH 3T3 cells.J Biol Chem. 1993 May 25;268(15):11456-62. J Biol Chem. 1993. PMID: 8496193
-
Involvement of carboxy-terminal amino acids in secretion of human lysosomal protease cathepsin L.Biochemistry. 1998 Jun 9;37(23):8584-94. doi: 10.1021/bi972251z. Biochemistry. 1998. PMID: 9622510
-
Protein determinants impair recognition of procathepsin L phosphorylated oligosaccharides by the cation-independent mannose 6-phosphate receptor.J Biol Chem. 1990 Jul 15;265(20):11864-71. J Biol Chem. 1990. PMID: 2164020
-
A shortcut to the lysosome: the mannose-6-phosphate-independent pathway.Mol Genet Metab. 2012 Nov;107(3):257-66. doi: 10.1016/j.ymgme.2012.07.012. Epub 2012 Jul 20. Mol Genet Metab. 2012. PMID: 22884962 Review.
-
Biosynthesis and alternate targeting of the lysosomal cysteine protease cathepsin L.Int Rev Cytol. 2004;241:1-51. doi: 10.1016/S0074-7696(04)41001-8. Int Rev Cytol. 2004. PMID: 15548418 Review.
Cited by
-
Procathepsin V Is Secreted in a TSH Regulated Manner from Human Thyroid Epithelial Cells and Is Accessible to an Activity-Based Probe.Int J Mol Sci. 2020 Nov 30;21(23):9140. doi: 10.3390/ijms21239140. Int J Mol Sci. 2020. PMID: 33266306 Free PMC article.
-
Proteolysis and antigen presentation by MHC class II molecules.Adv Immunol. 2002;80:71-114. doi: 10.1016/s0065-2776(02)80013-x. Adv Immunol. 2002. PMID: 12078484 Free PMC article. Review.
-
Exposure at the cell surface is required for gas3/PMP22 To regulate both cell death and cell spreading: implication for the Charcot-Marie-Tooth type 1A and Dejerine-Sottas diseases.Mol Biol Cell. 2000 Sep;11(9):2901-14. doi: 10.1091/mbc.11.9.2901. Mol Biol Cell. 2000. PMID: 10982389 Free PMC article.
-
Regulation of elastinolytic cysteine proteinase activity in normal and cathepsin K-deficient human macrophages.J Exp Med. 2000 Sep 18;192(6):789-99. doi: 10.1084/jem.192.6.789. J Exp Med. 2000. PMID: 10993910 Free PMC article.
References
-
- Achkar C, Gong Q, Frankfater A, Bajkowski AS. Differences in targeting and secretion of cathepsins B and L by BALB/3T3 fibroblasts and Moloney murine sarcoma virus-transformed BALB/3T3 fibroblasts. J Biol Chem. 1990;265:13650–13654. - PubMed
-
- Baker EN. Structure of actinidin, after refinement at 1.7 Å resolution. J Mol Biol. 1980;141:441–484. - PubMed
-
- Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The protein data bank: A computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. - PubMed
-
- Blundell TL, Sibanda BL, Sternberg MJE, Thorton JM. Knowledged-based prediction of protein structures and the design of novel molecules. Nature. 1987;326:347–352. - PubMed
-
- Boyer MJ, Tannock IF. Lysosomes, lysosomal enzymes, and cancer. Adv Cancer Res. 1993;60:269–291. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

