High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria
- PMID: 9572926
- PMCID: PMC106205
- DOI: 10.1128/AEM.64.5.1620-1627.1998
High levels of endemicity of 3-chlorobenzoate-degrading soil bacteria
Abstract
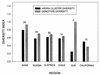
Soils samples were obtained from pristine ecosystems in six regions on five continents. Two of the regions were boreal forests, and the other four were Mediterranean ecosystems. Twenty-four soil samples from each of four or five sites in each of the regions were enriched by using 3-chlorobenzoate (3CBA), and 3CBA mineralizers were isolated from most samples. These isolates were analyzed for the ability to mineralize 3CBA, and genotypes were determined with repetitive extragenic palindromic PCR genomic fingerprints and restriction digests of the 16S rRNA genes (amplified ribosomal DNA restriction analysis [ARDRA]). We found that our collection of 150 stable 3CBA-mineralizing isolates included 48 genotypes and 44 ARDRA types, which formed seven distinct clusters. The majority (91%) of the genotypes were unique to the sites from which they were isolated, and each genotype was found only in the region from which it was isolated. A total of 43 of the 44 ARDRA types were found in only one region. A few genotypes were repeatedly found in one region but not in any other continental region, suggesting that they are regionally endemic. A correlation between bacterial genotype and vegetative community was found for the South African samples. These results suggest that the ability to mineralize 3CBA is distributed among very diverse genotypes and that the genotypes are not globally dispersed.
Figures



References
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1987.
-
- Brunsbach F R, Reineke W. Degradation of chlorobenzoates in soil slurry by special organisms. Appl Microbiol Biotechnol. 1993;39:117–122. - PubMed
-
- Chatterjee D K, Chakrabarty A M. Restriction mapping of a chlorobenzoate degradative plasmid and molecular cloning of the degradative genes. Gene. 1984;27:173–181. - PubMed
-
- de Bruijn F J. Use of repetitive (repetitive extragenic palindromic and enterobacterial repetitive intergeneric consensus) sequences and the polymerase chain reaction to fingerprint the genomes of Rhizobium meliloti isolates and other soil bacteria. Appl Environ Microbiol. 1992;58:2180–2187. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

