Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus
- PMID: 9572949
- PMCID: PMC106228
- DOI: 10.1128/AEM.64.5.1766-1772.1998
Molecular analysis of a laccase gene from the white rot fungus Pycnoporus cinnabarinus
Abstract
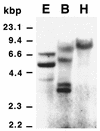
It was recently shown that the white rot basidiomycete Pycnoporus cinnabarinus secretes an unusual set of phenoloxidases when it is grown under conditions that stimulate ligninolysis (C. Eggert, U. Temp, and K.-E. L. Eriksson, Appl. Environ. Microbiol. 62:1151-1158, 1996). In this report we describe the results of a cloning and structural analysis of the laccase-encoding gene (lcc3-1) expressed by P. cinnabarinus during growth under xylidine-induced conditions. The coding region of the genomic laccase sequence, which is preceded by the eukaryotic promoter elements TATA and CAATA, spans more than 2,390 bp. The corresponding laccase cDNA was identical to the genomic sequence except for 10 introns that were 50 to 60 bp long. A sequence analysis indicated that the P. cinnabarinus lcc3-1 product has a Phe residue at a position likely to influence the reduction-oxidation potential of the enzyme's type 1 copper center. The P. cinnabarinus lcc3-1 sequence was most similar to the sequence encoding a laccase from Coriolus hirsutus (level of similarity, 84%).
Figures


References
-
- Ballance D J. Sequences important for gene expression in filamentous fungi. Yeast. 1986;2:229–236. - PubMed
-
- Bao W, O’Malley D M, Whetten R, Sederoff R R. A laccase associated with lignification in loblolly pine xylem. Science. 1993;260:672–674. - PubMed
-
- Bar-Nun N, Mayer A M. Cucurbitacins—repressors of induction of laccase formation. Phytochemistry. 1989;28:1369–1371.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

