Cloning and characterization of a prolinase gene (pepR) from Lactobacillus rhamnosus
- PMID: 9572959
- PMCID: PMC106238
- DOI: 10.1128/AEM.64.5.1831-1836.1998
Cloning and characterization of a prolinase gene (pepR) from Lactobacillus rhamnosus
Abstract
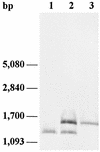
A peptidase gene expressing L-proline-beta-naphthylamide-hydrolyzing activity was cloned from a gene library of Lactobacillus rhamnosus 1/6 isolated from cheese. Peptidase-expressing activity was localized in a 1.5-kb SacI fragment. A sequence analysis of the SacI fragment revealed the presence of one complete open reading frame (ORF1) that was 903 nucleotides long. The ORF1-encoded 34.2-kDa protein exhibited 68% identity with the PepR protein from Lactobacillus helveticus. Additional sequencing revealed the presence of another open reading frame (ORF2) following pepR; this open reading frame was 459 bp long. Northern (RNA) and primer extension analyses indicated that pepR is expressed both as a monocistronic transcriptional unit and as a dicistronic transcriptional unit with ORF2. Gene replacement was used to construct a PepR-negative strain of L. rhamnosus. PepR was shown to be the primary enzyme capable of hydrolyzing Pro-Leu in L. rhamnosus. However, the PepR-negative mutant did not differ from the wild type in its ability to grow and produce acid in milk. The cloned pepR expressed activity against dipeptides with N-terminal proline residues. Also, Met-Ala, Leu-Leu, and Leu-Gly-Gly and the chromogenic substrates L-leucine-beta-naphthylamide and L-phenylalanine-beta-naphthylamide were hydrolyzed by the PepR of L. rhamnosus.
Figures


References
-
- Arora G, Lee B H. Purification and characterization of aminopeptidase from Lactobacillus casei subsp. casei LLG. J Dairy Sci. 1992;75:700–710.
-
- Arora G, Lee B H. Purification and characterization of an aminopeptidase from Lactobacillus casei subsp. rhamnosus S93. Biotechnol Appl Biochem. 1994;19:179–192. - PubMed
-
- Atlan D, Gilbert C, Blanc B, Portalier R. Cloning, sequencing and characterization of the pepIP gene encoding a proline iminopeptidase from Lactobacillus delbrueckii subsp. bulgaricus CNRZ 397. Microbiology. 1994;140:527–535. - PubMed
-
- Aymerich M T, Hugas M, Garriga M, Vogel R F, Monfort J M. Electrotransformation of meat lactobacilli. Effect of several parameters on their efficiency of transformation. J Appl Bacteriol. 1993;75:320–325.
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Miscellaneous

