Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose
- PMID: 9572962
- PMCID: PMC106241
- DOI: 10.1128/AEM.64.5.1852-1859.1998
Genetically engineered Saccharomyces yeast capable of effective cofermentation of glucose and xylose
Abstract
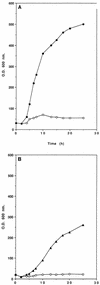
Xylose is one of the major fermentable sugars present in cellulosic biomass, second only to glucose. However, Saccharomyces spp., the best sugar-fermenting microorganisms, are not able to metabolize xylose. We developed recombinant plasmids that can transform Saccharomyces spp. into xylose-fermenting yeasts. These plasmids, designated pLNH31, -32, -33, and -34, are 2 microns-based high-copy-number yeast-E. coli shuttle plasmids. In addition to the geneticin resistance and ampicillin resistance genes that serve as dominant selectable markers, these plasmids also contain three xylose-metabolizing genes, a xylose reductase gene, a xylitol dehydrogenase gene (both from Pichia stipitis), and a xylulokinase gene (from Saccharomyces cerevisiae). These xylose-metabolizing genes were also fused to signals controlling gene expression from S. cerevisiae glycolytic genes. Transformation of Saccharomyces sp. strain 1400 with each of these plasmids resulted in the conversion of strain 1400 from a non-xylose-metabolizing yeast to a xylose-metabolizing yeast that can effectively ferment xylose to ethanol and also effectively utilizes xylose for aerobic growth. Furthermore, the resulting recombinant yeasts also have additional extraordinary properties. For example, the synthesis of the xylose-metabolizing enzymes directed by the cloned genes in these recombinant yeasts does not require the presence of xylose for induction, nor is the synthesis repressed by the presence of glucose in the medium. These properties make the recombinant yeasts able to efficiently ferment xylose to ethanol and also able to efficiently coferment glucose and xylose present in the same medium to ethanol simultaneously.
Figures




References
-
- Ammerer G. Expression of genes in yeast using the ADC1 promoter. Methods Enzymol. 1983;101:192–201. - PubMed
-
- Armstrong K A, Som T, Volkert F C, Rose A, Broach J R. Propagation and expression of genes in yeast using 2-micron circle vectors. In: Barr P J, Brake A J, Valenzuela P, editors. Yeast genetic engineering. Boston, Mass: Butterworths; 1989. pp. 165–192. - PubMed
-
- Becker D, Guarente L. High efficiency transformation of yeast by electroporation. Methods Enzymol. 1991;194:182–186. - PubMed
-
- Bennetzen J L, Hall B D. The primary structure of the Saccharomyces cerevisiae gene for alcohol dehydrogenase I. J Biol Chem. 1982;257:3018–3025. - PubMed
-
- Bolen P L, Detroy R W. Induction of NADPH-linked d-xylose reductase and NAD-linked xylitol dehydrogenase activities in Pachysolen tannophilus by d-xylose, l-arabinose, or d-galactose. Biotechnol Bioeng. 1985;27:302–307. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

