BXR, an embryonic orphan nuclear receptor activated by a novel class of endogenous benzoate metabolites
- PMID: 9573044
- PMCID: PMC316771
- DOI: 10.1101/gad.12.9.1269
BXR, an embryonic orphan nuclear receptor activated by a novel class of endogenous benzoate metabolites
Abstract
Nuclear receptors are ligand-modulated transcription factors that respond to steroids, retinoids, and thyroid hormones to control development and body physiology. Orphan nuclear receptors, which lack identified ligands, provide a unique, and largely untapped, resource to discover new principles of physiologic homeostasis. We describe the isolation and characterization of the vertebrate orphan receptor, BXR, which heterodimerizes with RXR and binds high-affinity DNA sites composed of a variant thyroid hormone response element. A bioactivity-guided screen of embryonic extracts revealed that BXR is activatable by low-molecular-weight molecules with spectral patterns distinct from known nuclear receptor ligands. Mass spectrometry and 1H NMR analysis identified alkyl esters of amino and hydroxy benzoic acids as potent, stereoselective activators. In vitro cofactor association studies, along with competable binding of radiolabeled compounds, establish these molecules as bona fide ligands. Benzoates comprise a new molecular class of nuclear receptor ligand and their activity suggests that BXR may control a previously unsuspected vertebrate signaling pathway.
Figures



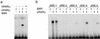
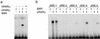




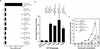
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
