The yeast Ada complex mediates the ligand-dependent activation function AF-2 of retinoid X and estrogen receptors
- PMID: 9573045
- PMCID: PMC316789
- DOI: 10.1101/gad.12.9.1278
The yeast Ada complex mediates the ligand-dependent activation function AF-2 of retinoid X and estrogen receptors
Abstract
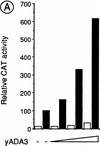
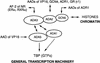
Nuclear receptors can function as ligand-inducible transregulators in both mammalian and yeast cells, indicating that important features of control of transcription have been conserved throughout evolution. Here, we report the isolation and characterization of a yeast protein that exhibits properties expected for a coactivator/mediator of the ligand-dependent activation function AF-2 present in the ligand-binding domain (LBD, region E) of the retinoid X (RXRalpha) and estrogen (ERalpha) receptors. This protein is identical to Ada3, a component of the yeast Ada coactivator complex. We demonstrate that: (1) the region encompassing residues 347-702 of Ada3 interacts with the LBD of RXRalpha and ERalpha in a ligand-dependent manner in yeast; (2) this interaction corresponds to a direct binding and requires the integrity of the core of the AF-2 activating domain (AF-2 AD) of both RXRalpha and ERalpha; (3) Ada3 as well as Ada2 and Gcn5, two other components of the Ada complex, are required for maximal AF-2 activity in yeast; and (4) Ada3 is able to enhance the AF-2 activity of RXRalpha and ERalpha when overexpressed in yeast and mammalian cells. Taken together, these data indicate that ligand-dependent transactivation by RXRalpha and ERalpha in yeast is mediated at least in part by the Ada complex, in which the Ada3 subunit directly binds to the holoreceptor LBD.
Figures








References
-
- Ayer DE, Lawrence QA, Eisenman RN. Mad–Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. - PubMed
-
- Baniahmad C, Nawaz Z, Baniahmad A, Gleeson MA, Tsai MJ, O’Malley BW. Enhancement of human estrogen receptor activity by SPT6: A potential coactivator. Mol Endocrinol. 1995;9:34–43. - PubMed
-
- Barlev NA, Candau R, Wang L, Darpino P, Silverman N, Berger SL. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. - PubMed
-
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
