Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor
- PMID: 9573046
- PMCID: PMC316766
- DOI: 10.1101/gad.12.9.1290
Control of distal antennal identity and tarsal development in Drosophila by spineless-aristapedia, a homolog of the mammalian dioxin receptor
Abstract
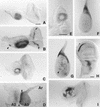
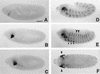
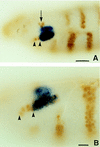
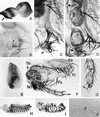
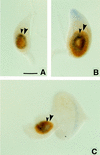
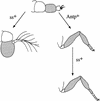
We report the molecular characterization of the spineless (ss) gene of Drosophila, and present evidence that it plays a central role in defining the distal regions of both the antenna and leg. ss encodes the closest known homolog of the mammalian dioxin receptor, a transcription factor of the bHLH-PAS family. Loss-of-function alleles of ss cause three major phenotypes: transformation of distal antenna to leg, deletion of distal leg (tarsal) structures, and reduction in size of most bristles. Consistent with these phenotypes, ss is expressed in the distal portion of the antennal imaginal disc, the tarsal region of each leg disc, and in bristle precursor cells. Ectopic expression of ss causes transformation of the maxillary palp and distal leg to distal antenna, and induces formation of an ectopic antenna in the rostral membrane. These effects indicate that ss plays a primary role in specifying distal antennal identity. In the tarsus, ss is expressed only early, and is required for later expression of the tarsal gene bric à brac (bab). Ectopic expression causes the deletion of medial leg structures, suggesting that ss plays an instructive role in the establishment of the tarsal primordium. In both the antenna and leg, ss expression is shown to depend on Distal-less (Dll), a master regulator of ventral appendage formation. The antennal transformation and tarsal deletions caused by ss loss-of-function mutations are probably atavistic, suggesting that ss played a central role in the evolution of distal structures in arthropod limbs.
Figures




References
-
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Baylies MK, Martinez Arias A, Bate M. wingless is required for the formation of a subset of muscle founder cells during Drosophila embryogenesis. Development. 1995;121:3829–3837. - PubMed
-
- Boudreaux HB. Arthropod Phylogeny, with special reference to insects. Malabar, FL: Robert E. Krieger Publishing; 1987.
-
- Brand A, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
