The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system
- PMID: 9573050
- PMCID: PMC316764
- DOI: 10.1101/gad.12.9.1338
The ClpXP and ClpAP proteases degrade proteins with carboxy-terminal peptide tails added by the SsrA-tagging system
Abstract
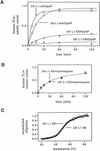
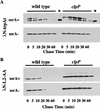
Interruption of translation in Escherichia coli can lead to the addition of an 11-residue carboxy-terminal peptide tail to the nascent chain. This modification is mediated by SsrA RNA (also called 10Sa RNA and tmRNA) and marks the tagged polypeptide for proteolysis. Degradation in vivo of lambda repressor amino-terminal domain variants bearing this carboxy-terminal SsrA peptide tag is shown here to depend on the cytoplasmic proteases ClpXP and ClpAP. Degradation in vitro of SsrA-tagged substrates was reproduced with purified components and required a substrate with a wild-type SsrA tail, the presence of both ClpP and either ClpA or ClpX, and ATP. Clp-dependent proteolysis accounts for most degradation of SsrA-tagged amino-domain substrates at 32 degrees C, but additional proteases contribute to the degradation of some of these SsrA-tagged substrates at 39 degrees C. The existence of multiple cytoplasmic proteases that function in SsrA quality-control surveillance suggests that the SsrA tag is designed to serve as a relatively promiscuous signal for proteolysis. Having diverse degradation systems able to recognize this tag may increase degradation capacity, permit degradation of a wide variety of different tagged proteins, or allow SsrA-tagged proteins to be degraded under different growth conditions.
Figures

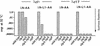




References
-
- Benyon RJ, Bond JS. Proteolytic enzymes: A practical approach. Oxford, UK: IRL Press; 1989.
-
- Bowie JU, Sauer RT. Identification of C-terminal extensions that protect proteins from intracellular proteolysis. J Biol Chem. 1989;264:7596–7602. - PubMed
-
- Clarke ND, Beamer LJ, Goldberg HR, Berkower C, Pabo CO. The DNA binding arm of λ repressor: Critical contacts from a flexible region. Science. 1991;254:267–270. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
