Suppression of crossing-over by DNA methylation in Ascobolus
- PMID: 9573054
- PMCID: PMC316785
- DOI: 10.1101/gad.12.9.1381
Suppression of crossing-over by DNA methylation in Ascobolus
Abstract
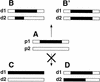
Homologous recombination between dispersed DNA repeats creates chromosomal rearrangements that are deleterious to the genome. The methylation associated with DNA repeats in many eukaryotes might serve to inhibit homologous recombination and play a role in preserving genome integrity. We have tested the hypothesis that DNA methylation suppresses meiotic recombination in the fungus Ascobolus immersus. The natural process of methylation-induced premeiotically (MIP) was used to methylate the b2 spore color gene, a 7.5-kb chromosomal recombination hot spot. The frequency of crossing-over between two markers flanking b2 was reduced several hundredfold when b2 was methylated on the two homologs. This demonstrates that DNA methylation strongly inhibits homologous recombination. When b2 was methylated on one homolog only, crossing-over was still reduced 50-fold, indicating that the effect of methylation cannot be limited to the blocking of initiation of recombination on the methylated homolog. On the basis of these and other observations, we propose that DNA methylation perturbs pairing between the two intact homologs before recombination initiation and/or impairs the normal processing of recombination intermediates.
Figures



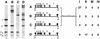


References
-
- Barlow DP. Methylation and imprinting: From host defense to gene regulation? Science. 1993;260:309–310. - PubMed
-
- Bennetzen JL, Schrick KS, Springer PS, Brown WE, SanMiguel P. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. - PubMed
-
- Bestor TH, Tycko B. Creation of genomic methylation patterns. Nature Genet. 1996;12:363–367. - PubMed
-
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for meiotic recombination, synaptonemal complex formation and cell cycle progression. Cell. 1992;69:439–456. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
