Integrated physical and genetic mapping of Bacillus cereus and other gram-positive bacteria based on IS231A transposition vectors
- PMID: 9573103
- PMCID: PMC108177
- DOI: 10.1128/IAI.66.5.2163-2169.1998
Integrated physical and genetic mapping of Bacillus cereus and other gram-positive bacteria based on IS231A transposition vectors
Abstract
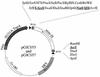
The genome structure of Bacillus cereus is relatively complex, its DNA being modulated between a size-varying chromosome and large plasmids. To study the genetic organization of the B. cereus type strain ATCC 14579, thermosensitive transposition vectors were designed on the basis of IS231A-derived cassettes containing uncommon restriction sites. A highly preferred insertion site for IS231A was detected in the chromosome by Southern blotting and pulsed-field gel electrophoresis (PFGE) analyses of independent insertion mutants. However, once this insertional hot spot was occupied, secondary IS231A insertions occurred randomly, as demonstrated by isolation of independent B. cereus auxotrophs at a frequency of approximately 0.6%. The hot-spot site, as well as several auxotrophic mutations, were mapped by using NotI, SfiI, and AscI PFGE restriction profiles. It was confirmed by sequencing that one of the insertions, generating an Ade- phenotype, had disrupted a gene of the purine synthesis pathway. These results showed that combined PFGE and sequencing analyses of mini-IS231A insertions enable the construction of integrated physical and genetic maps of B. cereus type strain. Moreover, the presence of the ultrarare I-SceI restriction site in the mini-IS231A allowed the isolation, in double-insertion mutants, of contiguous and nonoverlapping large chromosomal fragments, convenient for direct sequencing. The system detailed in this report is therefore a powerful tool for comparative genetic studies among members of the B. cereus group (i.e., B. cereus, B. thuringiensis, B. mycoides, and B. anthracis) and could also be applied to more distantly related gram-positive bacteria.
Figures



References
-
- Ash C, Farrow J A E, Dorsch M, Stackebrandt E, Collins M D. Comparative analysis of Bacillus anthracis, Bacillus cereus, and related species on the basis of reverse transcriptase sequencing of 16S rRNA. Int J Syst Bacteriol. 1991;41:343–346. - PubMed
-
- Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Short protocols in molecular biology. 3rd ed. New York, N.Y: John Wiley & Sons, Inc.; 1995. p. 2.11.
-
- Blattner F R, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. - PubMed
-
- Bloch C A, Rode C K, Obreque V H, Mahillon J. Purification of Escherichia coli chromosomal segments without cloning. Biochem Biophys Res Commun. 1996;223:104–111. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

