Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics
- PMID: 9573111
- PMCID: PMC108185
- DOI: 10.1128/IAI.66.5.2221-2229.1998
Use of a Mycobacterium tuberculosis H37Rv bacterial artificial chromosome library for genome mapping, sequencing, and comparative genomics
Abstract
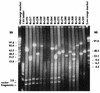
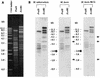
The bacterial artificial chromosome (BAC) cloning system is capable of stably propagating large, complex DNA inserts in Escherichia coli. As part of the Mycobacterium tuberculosis H37Rv genome sequencing project, a BAC library was constructed in the pBeloBAC11 vector and used for genome mapping, confirmation of sequence assembly, and sequencing. The library contains about 5,000 BAC clones, with inserts ranging in size from 25 to 104 kb, representing theoretically a 70-fold coverage of the M. tuberculosis genome (4.4 Mb). A total of 840 sequences from the T7 and SP6 termini of 420 BACs were determined and compared to those of a partial genomic database. These sequences showed excellent correlation between the estimated sizes and positions of the BAC clones and the sizes and positions of previously sequenced cosmids and the resulting contigs. Many BAC clones represent linking clones between sequenced cosmids, allowing full coverage of the H37Rv chromosome, and they are now being shotgun sequenced in the framework of the H37Rv sequencing project. Also, no chimeric, deleted, or rearranged BAC clones were detected, which was of major importance for the correct mapping and assembly of the H37Rv sequence. The minimal overlapping set contains 68 unique BAC clones and spans the whole H37Rv chromosome with the exception of a single gap of approximately 150 kb. As a postgenomic application, the canonical BAC set was used in a comparative study to reveal chromosomal polymorphisms between M. tuberculosis, M. bovis, and M. bovis BCG Pasteur, and a novel 12.7-kb segment present in M. tuberculosis but absent from M. bovis and M. bovis BCG was characterized. This region contains a set of genes whose products show low similarity to proteins involved in polysaccharide biosynthesis. The H37Rv BAC library therefore provides us with a powerful tool both for the generation and confirmation of sequence data as well as for comparative genomics and other postgenomic applications. It represents a major resource for present and future M. tuberculosis research projects.
Figures



References
-
- Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. - PubMed
-
- Bensimon A, Simon A, Chiffaudel A, Croquette V, Heslot F, Bensimon D. Alignment and sensitive detection of DNA by a moving interface. Science. 1994;265:2096–2098. - PubMed
-
- Bergh S, Cole S T. MycDB: an integrated mycobacterial database. Mol Microbiol. 1994;12:517–534. - PubMed
Publication types
MeSH terms
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

