The wzz (cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity
- PMID: 9573151
- PMCID: PMC107218
- DOI: 10.1128/JB.180.10.2670-2675.1998
The wzz (cld) protein in Escherichia coli: amino acid sequence variation determines O-antigen chain length specificity
Abstract
The O antigen is a polymer with a repeated unit. The chain length in most Escherichia coli strains has a modal value of 10 to 18 O units, but other strains have higher or lower modal values. wzz (cld/rol) mutants have a random chain length distribution, showing that the modal distribution is determined by the Wzz protein. Cloned wzz genes from E. coli strains with short (7 to 16), intermediate (10 to 18), and long (16 to 25) modal chain lengths were transferred to a model system, and their effects on O111 antigen were studied. The O111 chain length closely resembled that of the parent strains. We present data based on the construction of chimeric wzz genes and site-directed mutagenesis of the wzz gene to show that the modal value of O-antigen chain length of E. coli O1, O2, O7, and O157 strains can be changed by specific amino acid substitutions in wzz. It is concluded that the O-antigen chain length heterogeneity in E. coli strains is the result of amino acid sequence variation of the Wzz protein.
Figures


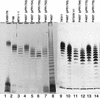
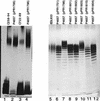


References
-
- Aucken H, Pitt T. Serological relationships of the O antigens of Klebsiella pneumoniae O5, Escherichia coli O8 and a new serotype of Serratia marcescens. FEMS Microbiol Lett. 1991;80:93–98. - PubMed
-
- Bastin D A, Brown P K, Haase A, Stevenson G, Reeves P R. Repeat unit polysaccharides of bacteria: a model for polymerisation resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–734. - PubMed
-
- Bastin D A, Romana L K, Reeves P R. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol Microbiol. 1991;5:2223–2231. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

