Isolation and characterization of Methanobacterium thermoautotrophicum DeltaH mutants unable to grow under hydrogen-deprived conditions
- PMID: 9573152
- PMCID: PMC107219
- DOI: 10.1128/JB.180.10.2676-2681.1998
Isolation and characterization of Methanobacterium thermoautotrophicum DeltaH mutants unable to grow under hydrogen-deprived conditions
Abstract
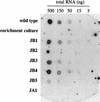
By using random mutagenesis and enrichment by chemostat culturing, we have developed mutants of Methanobacterium thermoautotrophicum that were unable to grow under hydrogen-deprived conditions. Physiological characterization showed that these mutants had poorer growth rates and growth yields than the wild-type strain. The mRNA levels of several key enzymes were lower than those in the wild-type strain. A fed-batch study showed that the expression levels were related to the hydrogen supply. In one mutant strain, expression of both methyl coenzyme M reductase isoenzyme I and coenzyme F420-dependent 5,10-methylenetetrahydromethanopterin dehydrogenase was impaired. The strain was also unable to form factor F390, lending support to the hypothesis that the factor functions in regulation of methanogenesis in response to changes in the availability of hydrogen.
Figures


References
-
- Bonacker L G, Baudner S, Thauer R K. Differential expression of the two methyl-coenzyme M reductases in Methanobacterium thermoautotrophicum as determined immunochemically via isoenzyme specific antisera. Eur J Biochem. 1992;206:87–92. - PubMed
-
- Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Roberts K M, Hurst M A, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. - PubMed
-
- Fardeau M-L, Belaich J-P. Energetics of the growth of Methanococcus thermolithotrophicus. Arch Microbiol. 1986;144:381–385.
-
- Gloss L M, Hausinger R P. Methanogen factor 390 formation: species distribution, reversibility and effects of non-oxidative cellular stress. BioFactors. 1988;1:237–240. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

