Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro
- PMID: 9573186
- PMCID: PMC107176
- DOI: 10.1128/JB.180.9.2359-2366.1998
Mutational analysis of the Chlamydia trachomatis rRNA P1 promoter defines four regions important for transcription in vitro
Abstract
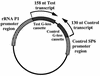
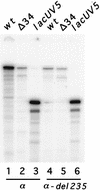
We have characterized the Chlamydia trachomatis ribosomal promoter, rRNA P1, by measuring the effect of substitutions and deletions on in vitro transcription with partially purified C. trachomatis RNA polymerase. Our analyses indicate that rRNA P1 contains potential -10 and -35 elements, analogous to Escherichia coli promoters recognized by E-sigma70. We identified a novel AT-rich region immediately downstream of the -35 region. The effect of this region was specific for C. trachomatis RNA polymerase and strongly attenuated by single G or C substitutions. Upstream of the -35 region was an AT-rich sequence that enhanced transcription by C. trachomatis and E. coli RNA polymerases. We propose that this region functions as an UP element.
Figures




Similar articles
-
Identification of sequences necessary for transcription in vitro from the Chlamydia trachomatis rRNA P1 promoter.J Bacteriol. 1996 Dec;178(23):6975-82. doi: 10.1128/jb.178.23.6975-6982.1996. J Bacteriol. 1996. PMID: 8955322 Free PMC article.
-
A positive cis-acting DNA element is required for high-level transcription in Chlamydia.J Bacteriol. 2000 Sep;182(18):5167-71. doi: 10.1128/JB.182.18.5167-5171.2000. J Bacteriol. 2000. PMID: 10960101 Free PMC article.
-
The RNA polymerase of Chlamydia trachomatis has a flexible sequence requirement at the -10 and -35 boxes of its promoters.J Bacteriol. 1994 Jun;176(12):3785-9. doi: 10.1128/jb.176.12.3785-3789.1994. J Bacteriol. 1994. PMID: 8206857 Free PMC article.
-
Strength and regulation without transcription factors: lessons from bacterial rRNA promoters.Cold Spring Harb Symp Quant Biol. 1998;63:131-9. doi: 10.1101/sqb.1998.63.131. Cold Spring Harb Symp Quant Biol. 1998. PMID: 10384277 Review. No abstract available.
-
Stringent control of bacterial transcription.Cell. 1985 May;41(1):6-8. doi: 10.1016/0092-8674(85)90050-9. Cell. 1985. PMID: 2581696 Review. No abstract available.
Cited by
-
Diversity of σ66-Specific Promoters Contributes to Regulation of Developmental Gene Expression in Chlamydia trachomatis.J Bacteriol. 2023 Jan 26;205(1):e0031022. doi: 10.1128/jb.00310-22. Epub 2023 Jan 4. J Bacteriol. 2023. PMID: 36598485 Free PMC article.
-
Arginine-dependent gene regulation via the ArgR repressor is species specific in chlamydia.J Bacteriol. 2006 Feb;188(3):919-27. doi: 10.1128/JB.188.3.919-927.2006. J Bacteriol. 2006. PMID: 16428395 Free PMC article.
-
Functional characterization of the principal sigma factor RpoD of phytoplasmas via an in vitro transcription assay.Sci Rep. 2015 Jul 7;5:11893. doi: 10.1038/srep11893. Sci Rep. 2015. PMID: 26150080 Free PMC article.
-
Characterization of in vitro DNA binding sites of the EUO protein of Chlamydia psittaci.Infect Immun. 2000 Mar;68(3):1337-49. doi: 10.1128/IAI.68.3.1337-1349.2000. Infect Immun. 2000. PMID: 10678946 Free PMC article.
-
Selective promoter recognition by chlamydial sigma28 holoenzyme.J Bacteriol. 2006 Nov;188(21):7364-77. doi: 10.1128/JB.01014-06. Epub 2006 Aug 25. J Bacteriol. 2006. PMID: 16936033 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

