Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator
- PMID: 9573203
- PMCID: PMC107193
- DOI: 10.1128/JB.180.9.2493-2501.1998
Regulation of benzoate degradation in Acinetobacter sp. strain ADP1 by BenM, a LysR-type transcriptional activator
Abstract
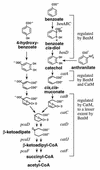
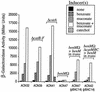
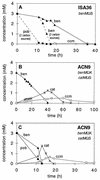
In Acinetobacter sp. strain ADP1, benzoate degradation requires the ben genes for converting benzoate to catechol and the cat genes for degrading catechol. Here we describe a novel transcriptional activator, BenM, that regulates the chromosomal ben and cat genes. BenM is homologous to CatM, a LysR-type transcriptional activator of the cat genes. Unusual regulatory features of this system include the abilities of both BenM and CatM to recognize the same inducer, cis,cis-muconate, and to regulate some of the same genes, such as catA and catB. Unlike CatM, BenM responded to benzoate. Benzoate together with cis,cis-muconate increased the BenM-dependent expression of the benABCDE operon synergistically. CatM was not required for this synergism, nor did CatM regulate the expression of a chromosomal benA::lacZ transcriptional fusion. BenM-mediated regulation differs significantly from that of the TOL plasmid-encoded conversion of benzoate to catechol in pseudomonads. The benM gene is immediately upstream of, and divergently transcribed from, benA, and a possible DNA binding site for BenM was identified between the two coding regions. Two mutations in the predicted operator/promoter region rendered ben gene expression either constitutive or inducible by cis,cis-muconate but not benzoate. Mutants lacking BenM, CatM, or both of these regulators degraded aromatic compounds at different rates, and the levels of intermediary metabolites that accumulated depended on the genetic background. These studies indicated that BenM is necessary for ben gene expression but not for expression of the cat genes, which can be regulated by CatM. In a catM-disrupted strain, BenM was able to induce higher levels of catA expression than catB expression.
Figures




References
-
- Bundy, B. M., and E. L. Neidle. Unpublished observation.
-
- Canovas J L, Stanier R Y. Regulation of the enzymes of the β-ketoadipate pathway in Moraxella calcoaceticus. Eur J Biochem. 1967;1:289–300. - PubMed
-
- Cebolla A, Sousa C, de Lorenzo V. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J Biol Chem. 1997;272:3986–3992. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

