The nucleoside triphosphatase and helicase activities of vaccinia virus NPH-II are essential for virus replication
- PMID: 9573237
- PMCID: PMC110003
- DOI: 10.1128/JVI.72.6.4729-4736.1998
The nucleoside triphosphatase and helicase activities of vaccinia virus NPH-II are essential for virus replication
Abstract
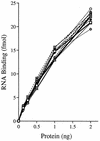
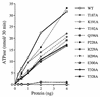
Vaccinia virus NPH-II is the prototypal RNA helicase of the DExH box protein family, which is defined by six shared sequence motifs. The contributions of conserved amino acids in motifs I (TGVGKTSQ), Ia (PRI), II (DExHE), and III (TAT) to enzyme activity were assessed by alanine scanning. NPH-II-Ala proteins were expressed in baculovirus-infected Sf9 cells, purified, and characterized with respect to their RNA helicase, nucleic acid-dependent ATPase, and RNA binding functions. Alanine substitutions at Lys-191 and Thr-192 (motif I), Arg-229 (motif Ia), and Glu-300 (motif II) caused severe defects in RNA unwinding that correlated with reduced rates of ATP hydrolysis. In contrast, alanine mutations at His-299 (motif II) and at Thr-326 and Thr-328 (motif III) elicited defects in RNA unwinding but spared the ATPase. None of the mutations analyzed affected the binding of NPH-II to RNA. These findings, together with previous mutational studies, indicate that NPH-II motifs I, Ia, II, and VI (QRxGRxGRxxxG) are essential for nucleoside triphosphate (NTP) hydrolysis, whereas motif III and the His moiety of the DExH-box serve to couple the NTPase and helicase activities. Wild-type and mutant NPH-II-Ala genes were tested for the ability to rescue temperature-sensitive nph2-ts viruses. NPH-II mutations that inactivated the phosphohydrolase in vitro were lethal in vivo, as judged by the failure to recover rescued viruses containing the Ala substitution. The NTPase activity was necessary, but not sufficient, to sustain virus replication, insofar as mutants for which NTPase was uncoupled from unwinding (H299A, T326A, and T328A) were also lethal. We conclude that the phosphohydrolase and helicase activities of NPH-II are essential for virus replication.
Figures

References
-
- Company M, Arenas J, Abelson J. Requirement of the RNA helicase-like protein PRP22 for release of messenger RNA from spliceosomes. Nature. 1991;349:487–493. - PubMed
-
- Fathi Z, Condit R C. Genetic and molecular biological characterization of a vaccinia virus temperature-sensitive complementation group affecting a virion component. Virology. 1991;181:258–272. - PubMed
-
- Fathi Z, Condit R C. Phenotypic characterization of a vaccinia virus temperature-sensitive complementation group affecting a virion component. Virology. 1991;181:273–276. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

