Cationic liposomes enhance the rate of transduction by a recombinant retroviral vector in vitro and in vivo
- PMID: 9573249
- PMCID: PMC110029
- DOI: 10.1128/JVI.72.6.4832-4840.1998
Cationic liposomes enhance the rate of transduction by a recombinant retroviral vector in vitro and in vivo
Abstract
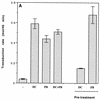
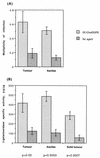
Cationic liposomes enhanced the rate of transduction of target cells with retroviral vectors. The greatest effect was seen with the formulation DC-Chol/DOPE, which gave a 20-fold increase in initial transduction rate. This allowed an efficiency of transduction after brief exposure of target cells to virus plus liposome that could be achieved only after extensive exposure to virus alone. Enhancement with DC-Chol/DOPE was optimal when stable virion-liposome complexes were preformed. The transduction rate for complexed virus, as for virus used alone or with the polycation Polybrene, showed first-order dependence on virus concentration. Cationic liposomes, but not Polybrene, were able to mediate envelope-independent transduction, but optimal efficiency required envelope-receptor interaction. When virus complexed with DC-Chol/DOPE was used to transduce human mesothelioma xenografts, transduction was enhanced four- to fivefold compared to that for virus alone. Since the efficacy of gene therapy is dependent on the number of cells modified, which is in turn dependent upon the balance between transduction and biological clearance of the vector, the ability of cationic liposomes to form stable complexes with retroviral vectors and enhance their rate of infection is likely to be important for in vivo application.
Figures





References
-
- Andersen K B. The fate of the surface protein gp70 during entry of retrovirus into mouse fibroblasts. Virology. 1985;142:112–120. - PubMed
-
- Andersen K B, Nexo B A. Entry of murine retrovirus into mouse fibroblasts. Virology. 1983;125:85–98. - PubMed
-
- Aubin R A, Weinfeld M, Mirzayans R, Paterson M C. Polybrene/DMSO-assisted gene transfer. Mol Biotechnol. 1994;1:29–48. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

