An evolutionarily conserved splice generates a secreted env-Bet fusion protein during human foamy virus infection
- PMID: 9573257
- PMCID: PMC110047
- DOI: 10.1128/JVI.72.6.4906-4910.1998
An evolutionarily conserved splice generates a secreted env-Bet fusion protein during human foamy virus infection
Abstract
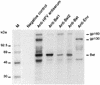
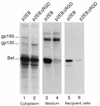
Foamy viruses (spumaretroviruses) represent a retroviral genus which exhibits unusual features relating it to pararetroviruses. Previously, we reported the existence of a protein species harboring Env, Bel, and Bet epitopes in human foamy virus (HFV)-infected cells (M. L. Giron, F. Rozain, M. C. Debons-Guillemin, M. Canivet, J. Périès, and R. Emanoil-Ravier, J. Virol. 67:3596-3600, 1993). Here, we identify this protein as a 160-kDa Env-Bet fusion glycoprotein (gp160) translated from an mRNA species harboring a highly conserved splice site which deletes the membrane anchor domain of Env and fuses the env open reading frame with that of bel1/bet. While gp160 and Bet proteins were both secreted into the supernatant, only Bet was taken up by recipient cells. Since Bet plays a key role in the switch from lytic to chronic infection, secretion of Bet and gp160, together with cellular uptake of Bet, could be highly relevant for both immune response and development of HFV infection in vivo.
Figures



Similar articles
-
Intra- and intercellular trafficking of the foamy virus auxiliary bet protein.J Virol. 2002 Apr;76(7):3388-94. doi: 10.1128/jvi.76.7.3388-3394.2002. J Virol. 2002. PMID: 11884565 Free PMC article.
-
Characterization of a human foamy virus 170-kilodalton Env-Bet fusion protein generated by alternative splicing.J Virol. 1998 May;72(5):4088-94. doi: 10.1128/JVI.72.5.4088-4094.1998. J Virol. 1998. PMID: 9557698 Free PMC article.
-
The human foamy virus internal promoter directs the expression of the functional Bel 1 transactivator and Bet protein early after infection.J Virol. 1994 Feb;68(2):638-45. doi: 10.1128/JVI.68.2.638-645.1994. J Virol. 1994. PMID: 8289367 Free PMC article.
-
[The research progress of foamy virus Bet protein].Bing Du Xue Bao. 2012 May;28(3):285-90. Bing Du Xue Bao. 2012. PMID: 22764533 Review. Chinese.
-
Non-primate foamy viruses.Curr Top Microbiol Immunol. 2003;277:197-211. doi: 10.1007/978-3-642-55701-9_9. Curr Top Microbiol Immunol. 2003. PMID: 12908774 Review.
Cited by
-
In vivo expression of a new hepatitis B virus protein encoded by a spliced RNA.J Clin Invest. 2000 Jan;105(1):55-60. doi: 10.1172/JCI8098. J Clin Invest. 2000. PMID: 10619861 Free PMC article.
-
Early steps of retrovirus replicative cycle.Retrovirology. 2004 May 14;1:9. doi: 10.1186/1742-4690-1-9. Retrovirology. 2004. PMID: 15169567 Free PMC article. Review.
-
Intra- and intercellular trafficking of the foamy virus auxiliary bet protein.J Virol. 2002 Apr;76(7):3388-94. doi: 10.1128/jvi.76.7.3388-3394.2002. J Virol. 2002. PMID: 11884565 Free PMC article.
-
Restriction of foamy viruses by APOBEC cytidine deaminases.J Virol. 2006 Jan;80(2):605-14. doi: 10.1128/JVI.80.2.605-614.2006. J Virol. 2006. PMID: 16378963 Free PMC article.
-
Foamy viruses are unconventional retroviruses.J Virol. 1999 Mar;73(3):1747-55. doi: 10.1128/JVI.73.3.1747-1755.1999. J Virol. 1999. PMID: 9971751 Free PMC article. Review. No abstract available.
References
-
- Chang K H, Day C, Walker J, Hyypia T, Stanway G. The nucleotide sequences of wild-type coxsackievirus A9 strains imply that an RGD motif in VP1 is functionally significant. J Gen Virol. 1992;73:621–626. - PubMed
-
- Cleveland D W, Fischer S G, Kirschner M W, Laemmli U K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1997;252:1102–1106. - PubMed
-
- D’Souza S E, Ginsberg M H, Plow E F. Arginyl-glycyl-aspartic acid (RGD): a cell adhesion motif. Trends Biochem Sci. 1991;16:246–250. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

