Computerized analysis of restriction fragment length polymorphism patterns: comparative evaluation of two commercial software packages
- PMID: 9574697
- PMCID: PMC104820
- DOI: 10.1128/JCM.36.5.1318-1323.1998
Computerized analysis of restriction fragment length polymorphism patterns: comparative evaluation of two commercial software packages
Abstract
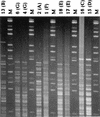
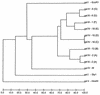
Two computerized restriction fragment length polymorphism pattern analysis systems, the BioImage system and the GelCompar system (Molecular Analyst Fingerprinting Plus in the United States), were compared. The two systems use different approaches to compare patterns from different gels. In GelCompar, a standard reference pattern in one gel is used to normalize subsequent gels containing lanes with the same reference pattern. In BioImage, the molecular sizes of the fragments are calculated from size standards present in each gel. The molecular size estimates obtained with the two systems for 12 restriction fragments of phage lambda were between 97 and 101% of their actual sizes, with a standard deviation of less than 1% of the average estimated size for most fragments. At the window sizes used for analysis, the GelCompar system performed somewhat better than BioImage in identifying visually identical patterns generated by electrophoretic separation of HhaI-restricted DNA of Listeria monocytogenes. Both systems require the user to make critical decisions in the analysis. It is very important to visually verify that the systems are finding all bands in each lane and that no artifacts are being detected; both systems allow manual editing. It is also important to verify results obtained in the pattern matching or clustering portions of the analysis.
Figures



Comment in
-
Critical observations on computerized analysis of banding patterns with commercial software packages.J Clin Microbiol. 1999 Mar;37(3):876-7. doi: 10.1128/JCM.37.3.876-877.1999. J Clin Microbiol. 1999. PMID: 10084889 Free PMC article. No abstract available.
References
-
- Dice L R. Measures of the amount of ecologic association between species. Ecology. 1945;26:297–302.
-
- Graves L M, Swaminathan B. Universal bacterial DNA isolation procedure. In: Persing D H, Smith T F, Tenover F C, White T J, editors. Diagnostic molecular microbiology: principles and applications. Washington, D.C: American Society for Microbiology; 1993. pp. 617–621.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

