Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus
- PMID: 9576772
- PMCID: PMC35019
- DOI: 10.1104/pp.117.1.33
Molecular characterization of the oxalate oxidase involved in the response of barley to the powdery mildew fungus
Abstract
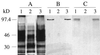
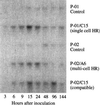
Previously we reported that oxalate oxidase activity increases in extracts of barley (Hordeum vulgare) leaves in response to the powdery mildew fungus (Blumeria [syn. Erysiphe] graminis f.sp. hordei) and proposed this as a source of H2O2 during plant-pathogen interactions. In this paper we show that the N terminus of the major pathogen-response oxalate oxidase has a high degree of sequence identity to previously characterized germin-like oxalate oxidases. Two cDNAs were isolated, pHvOxOa, which represents this major enzyme, and pHvOxOb', representing a closely related enzyme. Our data suggest the presence of only two oxalate oxidase genes in the barley genome, i.e. a gene encoding HvOxOa, which possibly exists in several copies, and a single-copy gene encoding HvOxOb. The use of 3' end gene-specific probes has allowed us to demonstrate that the HvOxOa transcript accumulates to 6 times the level of the HvOxOb transcript in response to the powdery mildew fungus. The transcripts were detected in both compatible and incompatible interactions with a similar accumulation pattern. The oxalate oxidase is found exclusively in the leaf mesophyll, where it is cell wall located. A model for a signal transduction pathway in which oxalate oxidase plays a central role is proposed for the regulation of the hypersensitive response.
Figures







References
-
- Anderson MLM, Young BD. Quantitative filter hybridisation. In: Hames BD, Higgins SJ, editors. Nucleic Acid Hybridisation—A Practical Approach. Oxford, UK: IRL Press; 1985. pp. 73–111.
-
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K (1987) Current Protocols in Molecular Biology 1987–1988. J Wiley & Sons, New York
-
- Baker JC, Orlandi EW. Active oxygen in plant pathogenesis. Annu Rev Phytopathol. 1995;33:299–321. - PubMed
-
- Berna A, Bernier F. Regulated expression of a wheat germin gene in tobacco: oxalate oxidase activity and apoplastic localization of the heterologous protein. Plant Mol Biol. 1997;33:417–429. - PubMed
-
- Bryngelsson T, Sommer-Knudsen J, Gregersen PL, Collinge DB, Ek B, Thordal-Christensen H. Purification, characterization and molecular cloning of basic PR-1-type pathogenesis-related proteins from barley. Mol Plant-Microbe Interact. 1994;7:267–275. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

